Pleasant Lake Example
This example is a simplified version of the Pleasant Lake model published by Fienen et al (2021). The goal of the Pleasant Lake model, part of the Central Sands Lake Study, was to address connections between groundwater abstraction and ecological function of a lake in central Wisconsin, USA (WDNR 2021; Figure 1). This required modeling at multiple scales. Fine discretization was needed near the lake for accurate simulation of water levels and groundwater-lake flux. A large model domain was also needed, to simulate farfield water-use activity (chiefly irrigated agriculture) in order to delineate a limit of connection, as well as to incorporate distant hydrologic boundaries. Adopting a fine enough discretization for the lake detail throughout the farfield would have resulted in a model with more cells than could be practically managed. To mitigate this, three models were combined: a large regional model built with MODFLOW-NWT (Niswonger et al. 2011), an intermediate MODFLOW 6 model inset within the regional model to simulate the irrigated agriculture area, and a refined MODFLOW 6 inset model (nested within the intermediate model) to simulate the lake. Regional groundwater flow and the effects of distant boundaries were simulated with the MODFLOW-NWT model, which was coupled sequentially (one-way) to the MODFLOW 6 models through time-varying specified head boundaries along the intermediate MODFLOW 6 model perimeter. The two MODFLOW 6 models were coupled dynamically (both ways) within the groundwater flow solution, allowing for feedback between the models. Estimates of groundwater recharge for the MODFLOW models were provided by a soil water balance code (SWB; Westenbroek et al 2018) simulation that could consider alternative assumptions of climate and land use. Net infiltration estimates from the SWB model were conveyed to the Recharge Packages in the MODFLOW models using the NetCDF functionality in Modflow-setup.
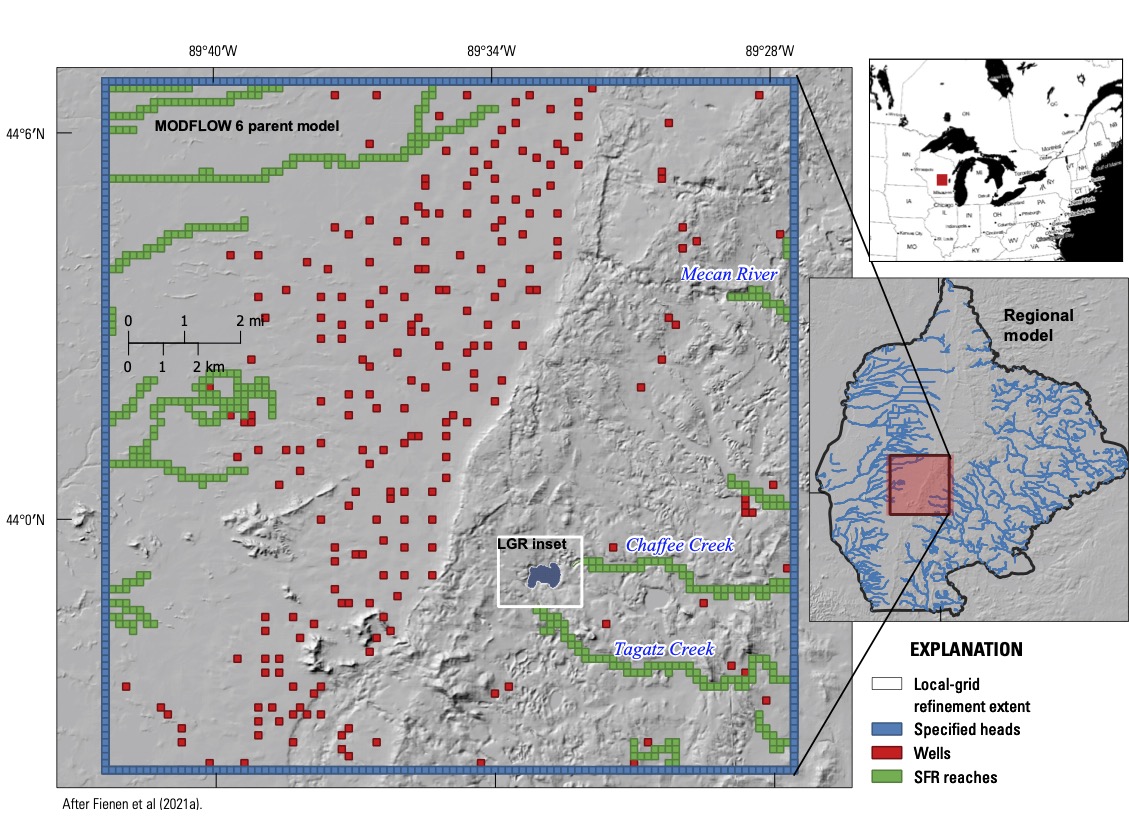
Figure 1: The full Pleasant Lake model domain with location map, showing the relationship between the regional, intermediate and LGR inset models, as well as the irrigation wells considered.
Most details of the Fienen et al (2021) model are included here, but to reduce file sizes and execution time, a smaller Modflow-6 simulation domain is used. The smaller domain is for illustration purposes only, and is not intended to be adequate for representing pumping impacts or providing a sufficient distance to the perimeter boundaries.
Model details
MODFLOW-6 simulation with a dynamically linked parent model and local grid refinement (LGR) inset model
LGR parent model is itself a Telescopic Mesh Refinment (TMR) inset from a regional MODFLOW-NWT model
Layer 1 in the regional model is subdivided evenly into two layers in the LGR models (
botm: from_parent: 0: -0.5). The other layers are mapped explicitly between the TMR parent and LGR models.starting heads for the LGR parent models were resampled from the regional model binary output
rch, npf, sto, and wel input copied from the regional model
SFR package constructed from an NHDPlus v2 dataset (path to NHDPlus files in the same structure as the
downloads from the NHDPlus website_)head observations from csv files with different column names
LGR inset extent based on a buffer distance around a feature of interest
LGR inset dis, ic, npf, sto and rch packages copied from LGR parent
Lake package created from polygon features, bathymetry raster, stage-area-volume file and climate data from
PRISM_.Lake package observations are set up automatically (output file for each lake)
[1]:
%%capture
import os
import numpy as np
import pandas as pd
from scipy.interpolate import griddata
import matplotlib.pyplot as plt
from matplotlib import patheffects
import flopy
import flopy.utils.binaryfile as bf
from mfsetup import MF6model
from mfsetup.discretization import cellids_to_kij
from gisutils import df2shp
import mfexport
from mfexport.utils import get_water_table
wd = os.getcwd()
Just make a model grid
Oftentimes at the start of a modeling project, we want to quickly test different grid resolutions and extents before attempting to build the model. We can do this with Modflow-setup by creating a model instance and then running the setup_grid() method. A model grid instance is created from the setup_grid: block in the configuration file.
Note: %%capture in the block below is just to suppress printing of stdout for display of this notebook in the modflow-setup documentation.
[2]:
%%capture
m = MF6model(cfg='pleasant_lgr_parent.yml')
m.setup_grid()
Since this model has local-grid refinement, it actually consists of two models: a parent built from pleasant_lgr_parent.yml, and an inset built from pleasant_lgr_inset.yml, which is referenced within pleasant_lgr_parent.yml. The two sub-models are connected and solved simulataneously within the same MODFLOW 6 simulation. A model grid is made for each sub-model. The model grids are instances of the MFsetupGrid grid class, a subclass of the Flopy StructuredGrid class with some
added functionality.
[3]:
m.modelgrid
[3]:
5 layer(s), 25 row(s), 25 column(s)
delr: [200.00...200.00] undefined
delc: [200.00...200.00] undefined
CRS: EPSG:3070
length units: meters
xll: 553400.0; yll: 387800.0; rotation: 0.0
Bounds: (np.float64(553400.0), np.float64(558400.0), np.float64(387800.0), np.float64(392800.0))
[4]:
m.inset['plsnt_lgr_inset'].modelgrid
[4]:
75 row(s), 85 column(s)
delr: [40.00...40.00] undefined
delc: [40.00...40.00] undefined
CRS: EPSG:3070
length units: meters
xll: 554200.0; yll: 388800.0; rotation: 0
Bounds: (np.float64(554200.0), np.float64(557600.0), np.float64(388800.0), np.float64(391800.0))
Working directory gottcha
Currently, to facilitate working with external files in Flopy, Modflow-setup changes the working directory to the model workspace. In the context of a flat script that only builds the model, this is fine, but in a notebook or other workflows, this can potentially cause confusion.
[5]:
os.getcwd()
[5]:
'/home/runner/work/modflow-setup/modflow-setup/examples/pleasant_lgr'
Write shapefiles of the inset and parent modelgrids
A shapefile of the grid bounding box is written by default on creation of the model grid, to the location specified by output_files: grid_file: in the setup_grid: block (default is <model workspace>/postproc/shps/). A shapefile of the grid cells as polygon features can be written as below:
[6]:
m.modelgrid.write_shapefile('postproc/shps/plsnt_lgr_parent_grid.shp')
m.inset['plsnt_lgr_inset'].modelgrid.write_shapefile('postproc/shps/plsnt_lgr_inset_grid.shp')
creating shapely Polygons of grid cells...
finished in 0.01s
writing postproc/shps/plsnt_lgr_parent_grid.shp... Done
creating shapely Polygons of grid cells...
finished in 0.19s
writing postproc/shps/plsnt_lgr_inset_grid.shp... Done
Change the working directory back to the notebook location
[7]:
os.chdir(wd)
Build the whole model
[8]:
%%capture
m = MF6model.setup_from_yaml('pleasant_lgr_parent.yml')
a MF6model instance (subclass of flopy.mf6.ModflowGwf) is returned
[9]:
m
[9]:
Pleasant Lake test case version 0.1.post58+g45e95f4
Parent model: /home/runner/work/modflow-setup/modflow-setup/examples/data/pleasant/pleasant
5 layer(s), 25 row(s), 25 column(s)
delr: [200.00...200.00] undefined
delc: [200.00...200.00] undefined
CRS: EPSG:3070
length units: meters
xll: 553400.0; yll: 387800.0; rotation: 0.0
Bounds: (np.float64(553400.0), np.float64(558400.0), np.float64(387800.0), np.float64(392800.0))
Packages: dis ic npf sto rcha_0 oc chd_0 obs_0 sfr_0 wel_0 obs_1 obs_2
13 period(s):
per start_datetime end_datetime perlen steady nstp
0 2012-01-01 2012-01-01 1.0 True 1
1 2012-01-01 2012-02-01 31.0 False 1
2 2012-02-01 2012-03-01 29.0 False 1
...
12 2012-12-01 2013-01-01 31.0 False 1
information from the configuration file is stored in an attached cfg dictionary
[10]:
m.cfg.keys()
[10]:
dict_keys(['metadata', 'simulation', 'model', 'parent', 'postprocessing', 'setup_grid', 'dis', 'tdis', 'ic', 'npf', 'sto', 'rch', 'sfr', 'high_k_lakes', 'lak', 'mvr', 'chd', 'drn', 'ghb', 'riv', 'wel', 'oc', 'obs', 'ims', 'mfsetup_options', 'filename', 'maw', 'external_files', 'intermediate_data', 'grid'])
the cfg dictionary contains both information from the configuration file, and MODFLOW input (such as external text file arrays) that was developed from the original source data. Internally in Modflow-setup, MODFLOW input in cfg is fed to the various Flopy object constructors.
[11]:
m.cfg['dis']
[11]:
defaultdict(dict,
{'remake_top': True,
'options': {'length_units': 'meters'},
'dimensions': {'nlay': 5, 'nrow': 25, 'ncol': 25},
'griddata': {'top': [{'filename': './external/plsnt_lgr_parent_top.dat'}],
'botm': [{'filename': './external/plsnt_lgr_parent_botm_000.dat'},
{'filename': './external/plsnt_lgr_parent_botm_001.dat'},
{'filename': './external/plsnt_lgr_parent_botm_002.dat'},
{'filename': './external/plsnt_lgr_parent_botm_003.dat'},
{'filename': './external/plsnt_lgr_parent_botm_004.dat'}],
'idomain': [{'filename': './external/plsnt_lgr_parent_idomain_000.dat'},
{'filename': './external/plsnt_lgr_parent_idomain_001.dat'},
{'filename': './external/plsnt_lgr_parent_idomain_002.dat'},
{'filename': './external/plsnt_lgr_parent_idomain_003.dat'},
{'filename': './external/plsnt_lgr_parent_idomain_004.dat'}]},
'top_filename_fmt': 'top.dat',
'botm_filename_fmt': 'botm_{:03d}.dat',
'idomain_filename_fmt': 'idomain_{:03d}.dat',
'minimum_layer_thickness': 1,
'drop_thin_cells': True,
'source_data': {'top': {'filename': '/home/runner/work/modflow-setup/modflow-setup/examples/data/pleasant/source_data/rasters/dem40m.tif',
'elevation_units': 'meters'},
'botm': {'filenames': {1: '/home/runner/work/modflow-setup/modflow-setup/examples/data/pleasant/source_data/rasters/botm0.tif',
2: '/home/runner/work/modflow-setup/modflow-setup/examples/data/pleasant/source_data/rasters/botm1.tif',
3: '/home/runner/work/modflow-setup/modflow-setup/examples/data/pleasant/source_data/rasters/botm2.tif',
4: '/home/runner/work/modflow-setup/modflow-setup/examples/data/pleasant/source_data/rasters/botm3.tif'}}},
'nlay': 4})
The inset LGR model is attached to the parent within an inset dictionary
[12]:
m.inset
[12]:
{'plsnt_lgr_inset': plsnt_lgr_inset model version 0.5.0.post58+g45e95f4
Parent model: ./plsnt_lgr_parent
5 layer(s), 75 row(s), 85 column(s)
delr: [40.00...40.00] undefined
delc: [40.00...40.00] undefined
CRS: EPSG:3070
length units: meters
xll: 554200.0; yll: 388800.0; rotation: 0
Bounds: (np.float64(554200.0), np.float64(557600.0), np.float64(388800.0), np.float64(391800.0))
Packages: dis ic npf sto rcha_0 oc sfr_0 lak_0 obs_0 obs_1
13 period(s):
per start_datetime end_datetime perlen steady nstp
0 2012-01-01 2012-01-01 1.0 True 1
1 2012-01-01 2012-02-01 31.0 False 1
2 2012-02-01 2012-03-01 29.0 False 1
...
12 2012-12-01 2013-01-01 31.0 False 1}
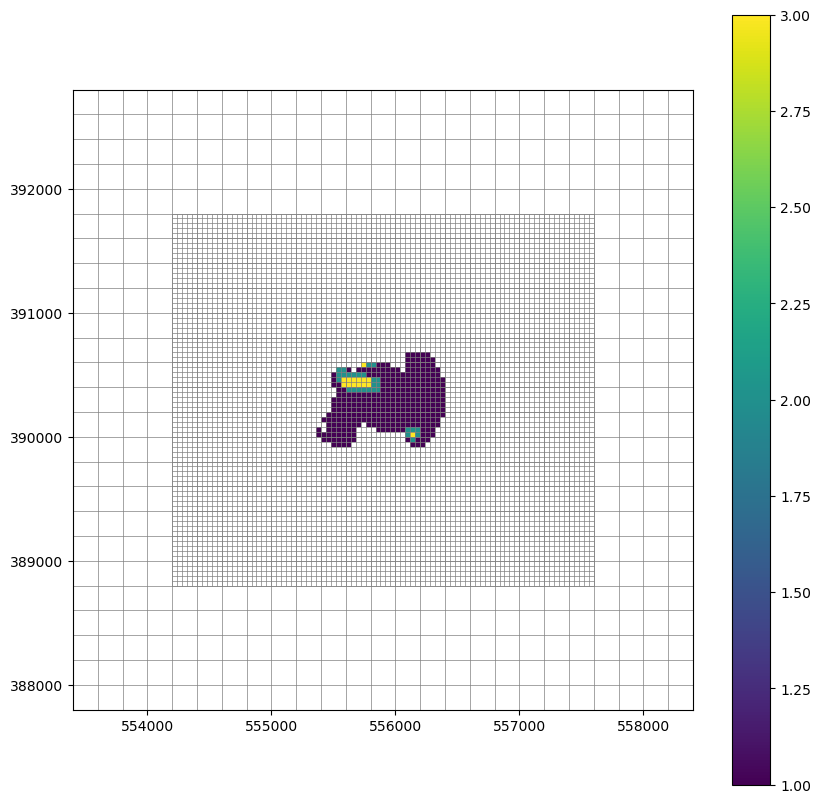
Plot the inset and parent model grids with Lake Package connections by layer
[13]:
inset = m.inset['plsnt_lgr_inset']
l, r, b, t = m.modelgrid.extent
layer = 0
fig, ax = plt.subplots(figsize=(10, 10))
parent_mv = flopy.plot.PlotMapView(model=m, ax=ax, layer=layer)
inset_mv = flopy.plot.PlotMapView(model=inset, ax=ax, layer=layer)
vconn = inset.lak.connectiondata.array[inset.lak.connectiondata.array['claktype'] == 'vertical']
k, i, j = cellids_to_kij(vconn['cellid'])
lakeconnections = np.zeros((inset.nrow, inset.ncol))
lakeconnections[i, j] = np.array(k+1)
lakeconnections = np.ma.masked_array(lakeconnections, mask=lakeconnections == 0)
qmi = inset_mv.plot_array(lakeconnections)
#inset_mv.plot_bc('LAK', color='navy')
#parent_mv.plot_bc('WEL_0', color='red')
lcp = parent_mv.plot_grid(lw=0.5, ax=ax)
lci = inset_mv.plot_grid(lw=0.5)
ax.set_ylim(b, t)
ax.set_xlim(l, r)
ax.set_aspect(1)
plt.colorbar(qmi)
[13]:
<matplotlib.colorbar.Colorbar at 0x7f45612fcf80>
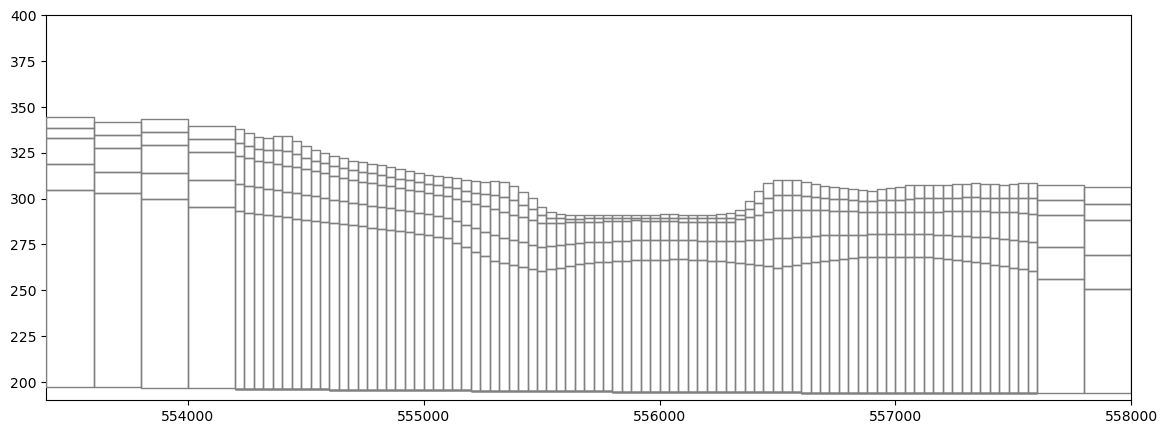
Make a cross section of the grid
[14]:
fig, ax = plt.subplots(figsize=(14, 5))
xs_line = [(553000, 390200), (558000, 390200)]
xs = flopy.plot.PlotCrossSection(model=m,
line={"line": xs_line}, ax=ax,
geographic_coords=True)
lc = xs.plot_grid(zorder=4)
xs2 = flopy.plot.PlotCrossSection(model=inset,
line={"line": xs_line}, ax=ax,
geographic_coords=True)
lc = xs2.plot_grid(zorder=4)
ax.set_ylim(190, 400)
plt.savefig('../../docs/source/_static/pleasant_lgr_xsection.png', bbox_inches='tight')
write the MODFLOW input files
(just like you would for a Flopy model)
[15]:
m.write_input()
writing simulation...
writing simulation name file...
writing simulation tdis package...
writing solution package ims_-1...
writing package pleasant_lgr.gwfgwf...
writing package pleasant_lgr.mvr...
writing model plsnt_lgr_parent...
writing model name file...
writing package dis...
writing package ic...
writing package npf...
writing package sto...
writing package rcha_0...
writing package oc...
writing package chd_0...
INFORMATION: maxbound in ('gwf6', 'chd', 'dimensions') changed to 269 based on size of stress_period_data
writing package obs_0...
writing package sfr_0...
writing package wel_0...
INFORMATION: maxbound in ('gwf6', 'wel', 'dimensions') changed to 2 based on size of stress_period_data
writing package obs_1...
writing package obs_2...
writing model plsnt_lgr_inset...
writing model name file...
writing package dis...
writing package ic...
writing package npf...
writing package sto...
writing package rcha_0...
writing package oc...
writing package sfr_0...
writing package lak_0...
writing package obs_0...
writing package obs_1...
SFRmaker v. 0.11.3.post35+g0f15d42
Running Flopy v. 3.9.0.dev2 diagnostics...
passed.
Checking for continuity in segment and reach numbering...
passed.
Checking for increasing segment numbers in downstream direction...
passed.
Checking for circular routing...
passed.
Checking reach connections for proximity...
2 segments with non-adjacent reaches found.
At segments:
1 2
2 segments with non-adjacent reaches found.
At segments:
1 2
Checking for model cells with multiple non-zero SFR conductances...
passed.
Checking for streambed tops of less than -10...
passed.
Checking for streambed tops of greater than 15000...
passed.
Checking segment_data for downstream rises in streambed elevation...
Segment elevup and elevdn not specified for nstrm=-16 and isfropt=1
passed.
Checking reach_data for downstream rises in streambed elevation...
7 reaches encountered with strtop < strtop of downstream reach.
Elevation rises:
k i j iseg ireach strtop strtopdn d_strtop reachID diff
4 20 15 2 1 290.66015625 292.6883850097656 2.028228759765625 5 -2.028228759765625
4 21 15 2 2 290.66015625 292.11285400390625 1.45269775390625 6 -1.45269775390625
2 24 20 2 10 288.81939697265625 290.1935119628906 1.374114990234375 14 -1.374114990234375
2 24 19 2 9 289.3415832519531 290.66015625 1.318572998046875 13 -1.318572998046875
2 24 18 2 8 289.4702453613281 290.66015625 1.189910888671875 12 -1.189910888671875
1 24 21 2 11 288.60980224609375 289.4702453613281 0.860443115234375 15 -0.860443115234375
4 23 18 2 7 290.1935119628906 290.66015625 0.466644287109375 11 -0.466644287109375
Checking reach_data for inconsistencies between streambed elevations and the model grid...
passed.
Checking segment_data for inconsistencies between segment end elevations and the model grid...
Segment elevup and elevdn not specified for nstrm=-16 and isfropt=1
passed.
Checking for streambed slopes of less than 0.0001...
passed.
Checking for streambed slopes of greater than 1.0...
passed.
wrote plsnt_lgr_parent_SFR.chk
wrote plsnt_lgr_parent.sfr.obs
converting reach and segment data to package data...
wrote /home/runner/work/modflow-setup/modflow-setup/examples/pleasant_lgr/external/plsnt_lgr_parent_packagedata.dat
wrote /home/runner/work/modflow-setup/modflow-setup/examples/pleasant_lgr/plsnt_lgr_parent.sfr
SFRmaker v. 0.11.3.post35+g0f15d42
Running Flopy v. 3.9.0.dev2 diagnostics...
passed.
Checking for continuity in segment and reach numbering...
passed.
Checking for increasing segment numbers in downstream direction...
passed.
Checking for circular routing...
passed.
Checking reach connections for proximity...
0 segments with non-adjacent reaches found.
At segments:
0 segments with non-adjacent reaches found.
At segments:
Checking for model cells with multiple non-zero SFR conductances...
1 model cells with multiple non-zero SFR conductances found.
This may lead to circular routing between collocated reaches.
Nodes with overlapping conductances:
k i j iseg ireach rchlen strthick strhc1
0 27 71 1 12 13.225701332092285 1.0 1.0
0 27 71 1 13 26.684703826904297 1.0 1.0
Checking for streambed tops of less than -10...
isfropt setting of 1,2 or 3 requires strtop information!
Checking for streambed tops of greater than 15000...
isfropt setting of 1,2 or 3 requires strtop information!
Checking segment_data for downstream rises in streambed elevation...
Segment elevup and elevdn not specified for nstrm=-41 and isfropt=1
passed.
Checking reach_data for downstream rises in streambed elevation...
passed.
Checking reach_data for inconsistencies between streambed elevations and the model grid...
passed.
Checking segment_data for inconsistencies between segment end elevations and the model grid...
Segment elevup and elevdn not specified for nstrm=-41 and isfropt=1
passed.
Checking for streambed slopes of less than 0.0001...
passed.
Checking for streambed slopes of greater than 1.0...
passed.
wrote plsnt_lgr_inset_SFR.chk
converting reach and segment data to package data...
wrote /home/runner/work/modflow-setup/modflow-setup/examples/pleasant_lgr/external/plsnt_lgr_inset_packagedata.dat
wrote /home/runner/work/modflow-setup/modflow-setup/examples/pleasant_lgr/plsnt_lgr_inset.sfr
Run the model
Note: Running the model through Flopy (as below) requires specification of the MODFLOW executable. In Flopy, the executable is specified via the exe_name argument to the simulation constructor for MODFLOW 6, or model constructor for previous MODFLOW versions. Similarly, in Modflow-setup, the exe_name is specified in the simulation: or model: block of the configuration file.
This example assumes that a MODFLOW 6 executable with the name “mf6” either resides in the model workspace, or is included in the system path.
[16]:
m.simulation.run_simulation()
FloPy is using the following executable to run the model: ../../../../../.local/bin/mf6
MODFLOW 6
U.S. GEOLOGICAL SURVEY MODULAR HYDROLOGIC MODEL
VERSION 6.3.0 release candidate 03/08/2022
***DEVELOP MODE***
MODFLOW 6 compiled Mar 08 2022 20:13:10 with Intel(R) Fortran Intel(R) 64
Compiler Classic for applications running on Intel(R) 64, Version 2021.5.0
Build 20211109_000000
This software is preliminary or provisional and is subject to
revision. It is being provided to meet the need for timely best
science. The software has not received final approval by the U.S.
Geological Survey (USGS). No warranty, expressed or implied, is made
by the USGS or the U.S. Government as to the functionality of the
software and related material nor shall the fact of release
constitute any such warranty. The software is provided on the
condition that neither the USGS nor the U.S. Government shall be held
liable for any damages resulting from the authorized or unauthorized
use of the software.
Run start date and time (yyyy/mm/dd hh:mm:ss): 2024/11/14 14:20:06
Writing simulation list file: mfsim.lst
Using Simulation name file: mfsim.nam
Solving: Stress period: 1 Time step: 1
Solving: Stress period: 2 Time step: 1
Solving: Stress period: 3 Time step: 1
Solving: Stress period: 4 Time step: 1
Solving: Stress period: 5 Time step: 1
Solving: Stress period: 6 Time step: 1
Solving: Stress period: 7 Time step: 1
Solving: Stress period: 8 Time step: 1
Solving: Stress period: 9 Time step: 1
Solving: Stress period: 10 Time step: 1
Solving: Stress period: 11 Time step: 1
Solving: Stress period: 12 Time step: 1
Solving: Stress period: 13 Time step: 1
Run end date and time (yyyy/mm/dd hh:mm:ss): 2024/11/14 14:20:10
Elapsed run time: 4.163 Seconds
Normal termination of simulation.
[16]:
(True, [])
Plot the head results
[17]:
tmr_parent_headsobj = bf.HeadFile('../data/pleasant/pleasant.hds')
lgr_parent_headsobj = bf.HeadFile('plsnt_lgr_parent.hds')
lgr_inset_headsobj = bf.HeadFile('plsnt_lgr_inset.hds')
# read the head results for the last stress period
kper = 12
lgr_parent_hds = lgr_parent_headsobj.get_data(kstpkper=(0, kper))
lgr_inset_hds = lgr_inset_headsobj.get_data(kstpkper=(0, kper))
# Get the water table elevation from the 3D head results
inset_wt = get_water_table(lgr_inset_hds, nodata=1e30)
parent_wt = get_water_table(lgr_parent_hds, nodata=1e30)
# put in the lake level (not included in head output)
lake_results = pd.read_csv('lake1.obs.csv')
stage = lake_results['STAGE'][kper]
inset_wt[inset.lakarr[0] == 1] = stage
First combine the parent and inset model head results
(into a single grid at the inset model resolution; for a nicer looking plot)
[18]:
# make the single grid
l, b, r, t = m.modelgrid.bounds
xi = np.arange(l, r, 40)
yi = np.arange(b, t, 40)[::-1]
Xi, Yi = np.meshgrid(xi, yi)
# make a single set of points
# including both parent and inset cell centers
# and water table values
x = m.modelgrid.xcellcenters[~parent_wt.mask]
y = m.modelgrid.ycellcenters[~parent_wt.mask]
x = np.append(x, inset.modelgrid.xcellcenters[~inset_wt.mask])
y = np.append(y, inset.modelgrid.ycellcenters[~inset_wt.mask])
z = parent_wt[~parent_wt.mask].data
z = np.append(z, inset_wt[~inset_wt.mask].data)
# interpolate the results from the points
# onto the single inset resolution grid
results = griddata((x, y), z, (Xi, Yi))
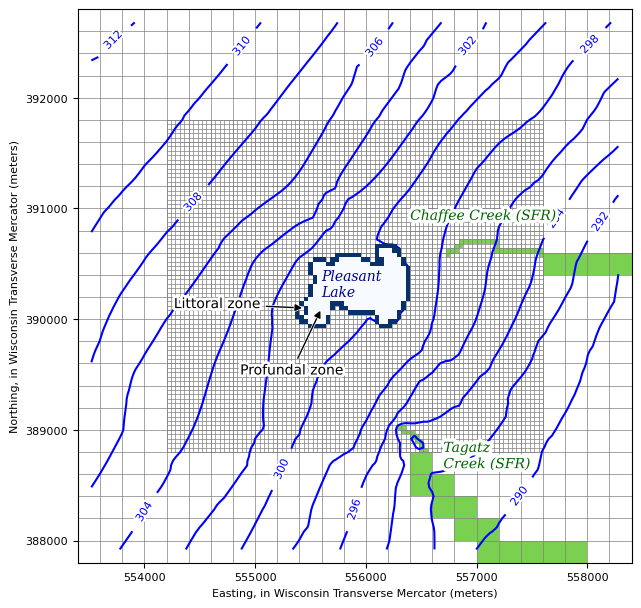
Make the plot
include the parent and inset model grids
show the head contours for the combined parent/inset simulation
show SFR boundary condition cells in green
show the lakebed leakance zones
[19]:
plt.rcParams['axes.labelsize'] = 8
plt.rcParams['xtick.labelsize'] = 8
plt.rcParams['ytick.labelsize'] = 8
layer = 0
fig, ax = plt.subplots(figsize=(6.5, 6.5))
# create Flopy plot objects
parent_mv = flopy.plot.PlotMapView(model=m, ax=ax, layer=layer)
inset_mv = flopy.plot.PlotMapView(model=inset, ax=ax, layer=layer)
# plot boundary condition cells from Modflow-setup array properties
inset_bcs = np.ma.masked_array(inset._isbc2d, mask=inset._isbc2d==0)
parent_bcs = np.ma.masked_array(m._isbc2d, mask=m._isbc2d==0)
parent_mv.plot_array(parent_bcs, vmin=0, vmax=5)
inset_mv.plot_array(inset_bcs, vmin=0, vmax=5)
#bdlknc_values = inset.lak.connectiondata.array['bedleak']
conn = inset.lak.connectiondata.array
k, i, j = cellids_to_kij(conn['cellid'])
bdlknc = np.zeros((inset.nlay, inset.nrow, inset.ncol))
bdlknc[k, i, j] = conn['bedleak']
bdlknc = np.max(bdlknc, axis=0)
bdlknc = np.ma.masked_array(bdlknc, mask=bdlknc == 0)
inset_mv.plot_array(bdlknc, cmap='Blues', zorder=200)
# contour the combined inset/parent head results
levels = np.arange(290, 315, 2)
ctr = ax.contour(Xi, Yi, results, levels=levels, colors='b', zorder=10)
labels = ax.clabel(ctr, inline=True, fontsize=8, inline_spacing=10)
plt.setp(labels, path_effects=[
patheffects.withStroke(linewidth=3, foreground="w")])
# plot the grid cell edges
lcp = parent_mv.plot_grid(lw=0.5, ax=ax)
lci = inset_mv.plot_grid(lw=0.5)
ax.set_ylim(b, t)
ax.set_xlim(l, r)
ax.set_aspect(1)
ax.set_ylabel('Northing, in Wisconsin Transverse Mercator (meters)')
ax.set_xlabel('Easting, in Wisconsin Transverse Mercator (meters)')
ax.text(555600, 390450, 'Pleasant\nLake', ha='left', va='top', color='DarkBlue',
fontsize=10, fontstyle='italic', family='Serif', zorder=202)
txt = ax.text(556400, 391000, 'Chaffee Creek (SFR)', ha='left', va='top', color='DarkGreen',
fontsize=10, fontstyle='italic', family='Serif', zorder=20)
txt.set_path_effects([patheffects.withStroke(linewidth=5, foreground='w')])
txt = ax.text(556700, 388900, 'Tagatz\nCreek (SFR)', ha='left', va='top', color='DarkGreen',
fontsize=10, fontstyle='italic', family='Serif', zorder=20)
txt.set_path_effects([patheffects.withStroke(linewidth=5, foreground='w')])
txt = ax.annotate("Littoral zone",
xy=(555450, 390100), xycoords='data',
xytext=(555050,390100), textcoords='data',
ha='right',
arrowprops=dict(arrowstyle="-|>",
connectionstyle="arc3", fc='k'),
path_effects=[patheffects.withStroke(linewidth=4, foreground='w')],
zorder=203
)
txt.arrow_patch.set_path_effects([
patheffects.Stroke(linewidth=2, foreground="w"),
patheffects.Normal()])
txt = ax.annotate("Profundal zone",
xy=(555600, 390100), xycoords='data',
xytext=(555800,389500), textcoords='data',
ha='right',
arrowprops=dict(arrowstyle="-|>",
connectionstyle="arc3", fc='k'),
path_effects=[patheffects.withStroke(linewidth=4, foreground='w')],
zorder=203
)
txt.arrow_patch.set_path_effects([
patheffects.Stroke(linewidth=2, foreground="w"),
patheffects.Normal()])
plt.tight_layout()
plt.savefig('postproc/pdfs/figure_2.pdf')
Use Modflow-export to export the modflow input to PDFs, rasters and shapefiles
[20]:
for model in m, inset:
mfexport.export(model, model.modelgrid, output_path=f'postproc/{model.name}/')
creating postproc/plsnt_lgr_parent/pdfs...
creating postproc/plsnt_lgr_parent/shps...
creating postproc/plsnt_lgr_parent/rasters...
dis package...
wrote postproc/plsnt_lgr_parent/rasters/thickness_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/thickness_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/thickness_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/thickness_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/thickness_lay4.tif
top:
wrote postproc/plsnt_lgr_parent/rasters/top.tif
botm:
wrote postproc/plsnt_lgr_parent/rasters/botm_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/botm_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/botm_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/botm_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/botm_lay4.tif
idomain:
wrote postproc/plsnt_lgr_parent/rasters/idomain_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/idomain_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/idomain_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/idomain_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/idomain_lay4.tif
ic package...
strt:
wrote postproc/plsnt_lgr_parent/rasters/strt_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/strt_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/strt_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/strt_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/strt_lay4.tif
npf package...
icelltype:
wrote postproc/plsnt_lgr_parent/rasters/icelltype_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/icelltype_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/icelltype_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/icelltype_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/icelltype_lay4.tif
k:
wrote postproc/plsnt_lgr_parent/rasters/k_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/k_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/k_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/k_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/k_lay4.tif
k33:
wrote postproc/plsnt_lgr_parent/rasters/k33_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/k33_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/k33_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/k33_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/k33_lay4.tif
sto package...
iconvert:
wrote postproc/plsnt_lgr_parent/rasters/iconvert_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/iconvert_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/iconvert_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/iconvert_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/iconvert_lay4.tif
ss:
wrote postproc/plsnt_lgr_parent/rasters/ss_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/ss_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/ss_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/ss_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/ss_lay4.tif
sy:
wrote postproc/plsnt_lgr_parent/rasters/sy_lay0.tif
wrote postproc/plsnt_lgr_parent/rasters/sy_lay1.tif
wrote postproc/plsnt_lgr_parent/rasters/sy_lay2.tif
wrote postproc/plsnt_lgr_parent/rasters/sy_lay3.tif
wrote postproc/plsnt_lgr_parent/rasters/sy_lay4.tif
rcha_0 package...
irch:
wrote postproc/plsnt_lgr_parent/rasters/irch_per0.tif
recharge:
wrote postproc/plsnt_lgr_parent/rasters/recharge_per0.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per1.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per2.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per3.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per4.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per5.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per6.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per7.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per8.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per9.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per10.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per11.tif
wrote postproc/plsnt_lgr_parent/rasters/recharge_per12.tif
chd_0 package...
writing postproc/plsnt_lgr_parent/shps/chd0_stress_period_data.shp... Done
head:
boundname:
Warning, variable: boundname
Export of non-period data from transientlists not implemented!
obs_0 package...
skipped, not implemented yet
sfr_0 package...
writing postproc/plsnt_lgr_parent/shps/plsnt_lgr_parent.sfr.shp... Done
wel_0 package...
writing postproc/plsnt_lgr_parent/shps/wel0_stress_period_data.shp... Done
q:
boundname:
Warning, variable: boundname
Export of non-period data from transientlists not implemented!
obs_1 package...
skipped, not implemented yet
obs_2 package...
writing postproc/plsnt_lgr_parent/shps/obs2_stress_period_data.shp... Done
creating postproc/plsnt_lgr_inset/pdfs...
creating postproc/plsnt_lgr_inset/shps...
creating postproc/plsnt_lgr_inset/rasters...
dis package...
wrote postproc/plsnt_lgr_inset/rasters/thickness_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/thickness_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/thickness_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/thickness_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/thickness_lay4.tif
top:
wrote postproc/plsnt_lgr_inset/rasters/top.tif
botm:
wrote postproc/plsnt_lgr_inset/rasters/botm_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/botm_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/botm_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/botm_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/botm_lay4.tif
idomain:
wrote postproc/plsnt_lgr_inset/rasters/idomain_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/idomain_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/idomain_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/idomain_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/idomain_lay4.tif
ic package...
strt:
wrote postproc/plsnt_lgr_inset/rasters/strt_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/strt_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/strt_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/strt_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/strt_lay4.tif
npf package...
icelltype:
wrote postproc/plsnt_lgr_inset/rasters/icelltype_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/icelltype_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/icelltype_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/icelltype_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/icelltype_lay4.tif
k:
wrote postproc/plsnt_lgr_inset/rasters/k_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/k_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/k_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/k_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/k_lay4.tif
k33:
wrote postproc/plsnt_lgr_inset/rasters/k33_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/k33_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/k33_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/k33_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/k33_lay4.tif
sto package...
iconvert:
wrote postproc/plsnt_lgr_inset/rasters/iconvert_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/iconvert_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/iconvert_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/iconvert_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/iconvert_lay4.tif
ss:
wrote postproc/plsnt_lgr_inset/rasters/ss_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/ss_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/ss_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/ss_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/ss_lay4.tif
sy:
wrote postproc/plsnt_lgr_inset/rasters/sy_lay0.tif
wrote postproc/plsnt_lgr_inset/rasters/sy_lay1.tif
wrote postproc/plsnt_lgr_inset/rasters/sy_lay2.tif
wrote postproc/plsnt_lgr_inset/rasters/sy_lay3.tif
wrote postproc/plsnt_lgr_inset/rasters/sy_lay4.tif
rcha_0 package...
irch:
wrote postproc/plsnt_lgr_inset/rasters/irch_per0.tif
recharge:
wrote postproc/plsnt_lgr_inset/rasters/recharge_per0.tif
sfr_0 package...
writing postproc/plsnt_lgr_inset/shps/plsnt_lgr_inset.sfr.shp... Done
lak_0 package...
skipping lak0.perioddata; efficient export not implemented
obs_0 package...
skipped, not implemented yet
obs_1 package...
writing postproc/plsnt_lgr_inset/shps/obs1_stress_period_data.shp... Done
Modflow-export can also create a summary table of the model inputs
[21]:
for model in m, inset:
mfexport.summarize(model, output_path=f'postproc/{model.name}/')
summarizing plsnt_lgr_parent input...
skipped, not implemented yet
skipped, not implemented yet
summarizing plsnt_lgr_inset input...
skipped, not implemented yet