USGS Water Quality Data
Introduction to dataRetrieval
Introduction
In this ~90 minute introduction, the goal is:
Introduce the modern
dataRetrievalworkflows.The intended audience is someone:
New to
dataRetrievalHas some R experience
RStudio Orientation
By default will look like:
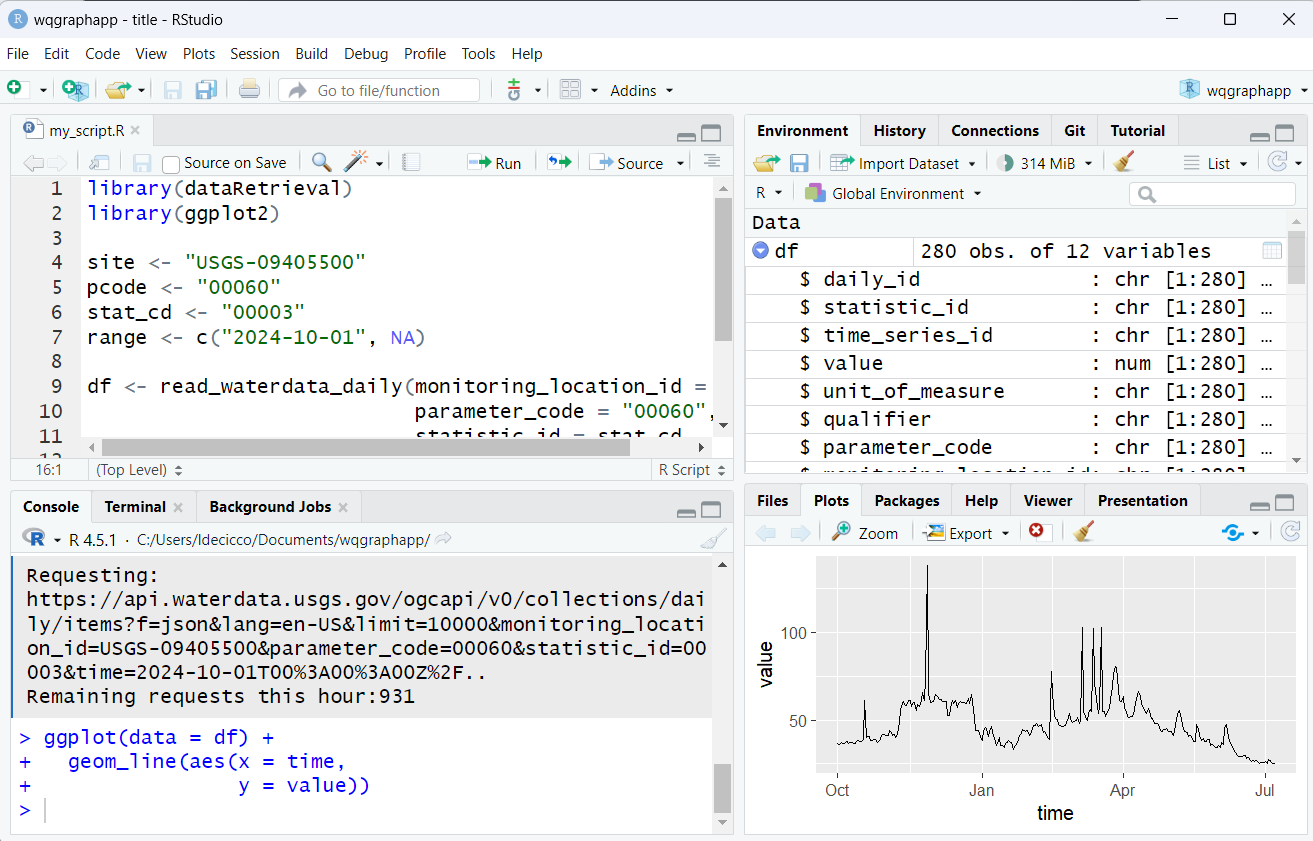
RStudio Appearances
Go to Tools -> Global Options -> Appearances to change style.
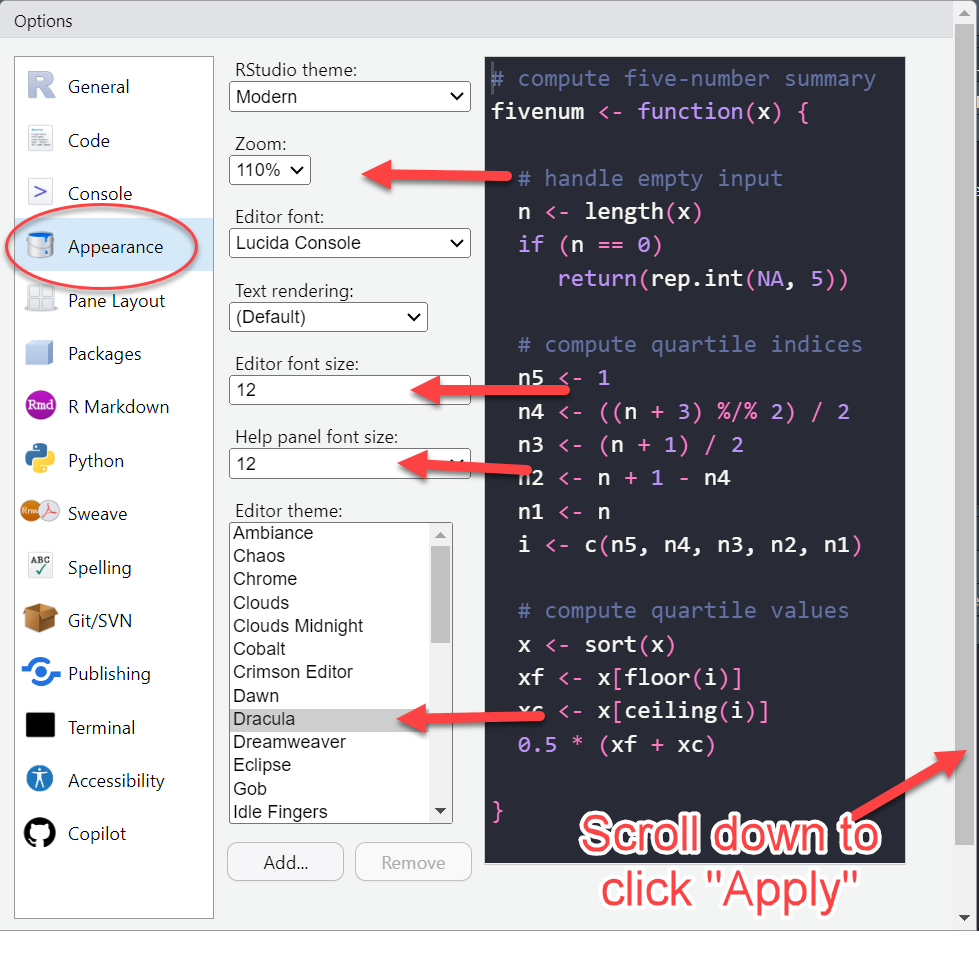
RStudio Orientation
Create scripts.
See code run.
See what variables are loaded
- Click on a data frame to View
- Plots and more

dataRetrieval: R-package for US water data
USGS Water Data APIs *
Surface water levels
Groundwater levels
Site metadata
Peak flows
Rating curves
Discrete water-quality data
Water Quality Portal (WQP) Data
Discrete water-quality data
USGS and non-USGS data
Installation
dataRetrieval is available on the Comprehensive R Archive Network (CRAN) repository. To install dataRetrieval on your computer, open RStudio and run this line of code in the Console:
Then each time you open R, you’ll need to load the library:
dataRetrieval: External Documentation
dataRetrieval: External Documentation
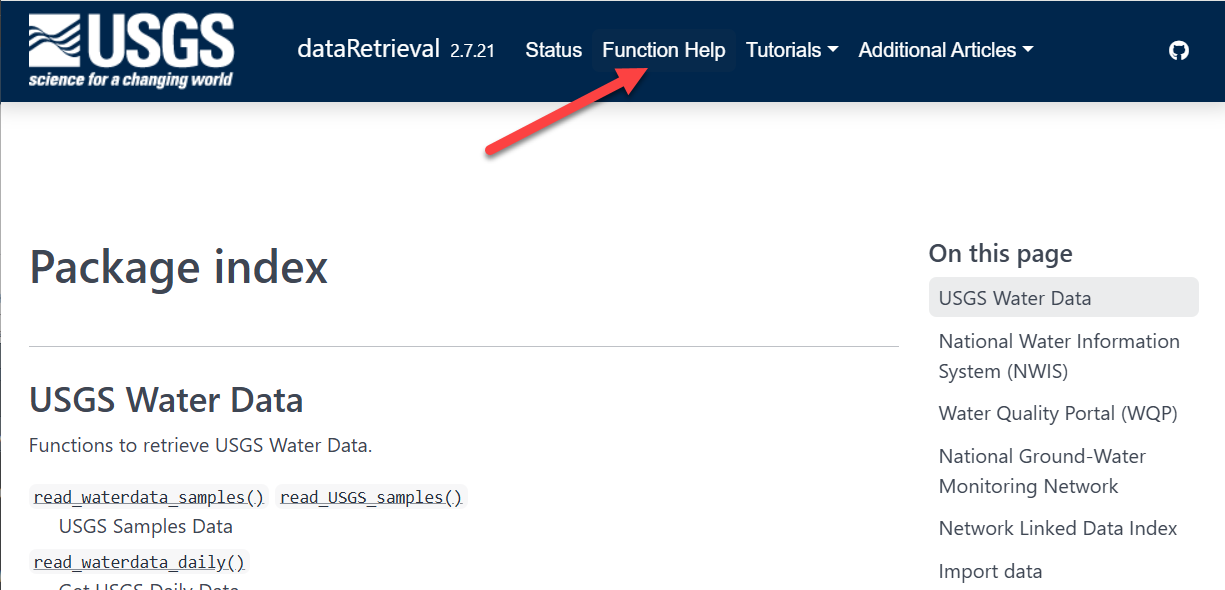
dataRetrieval: External Documentation
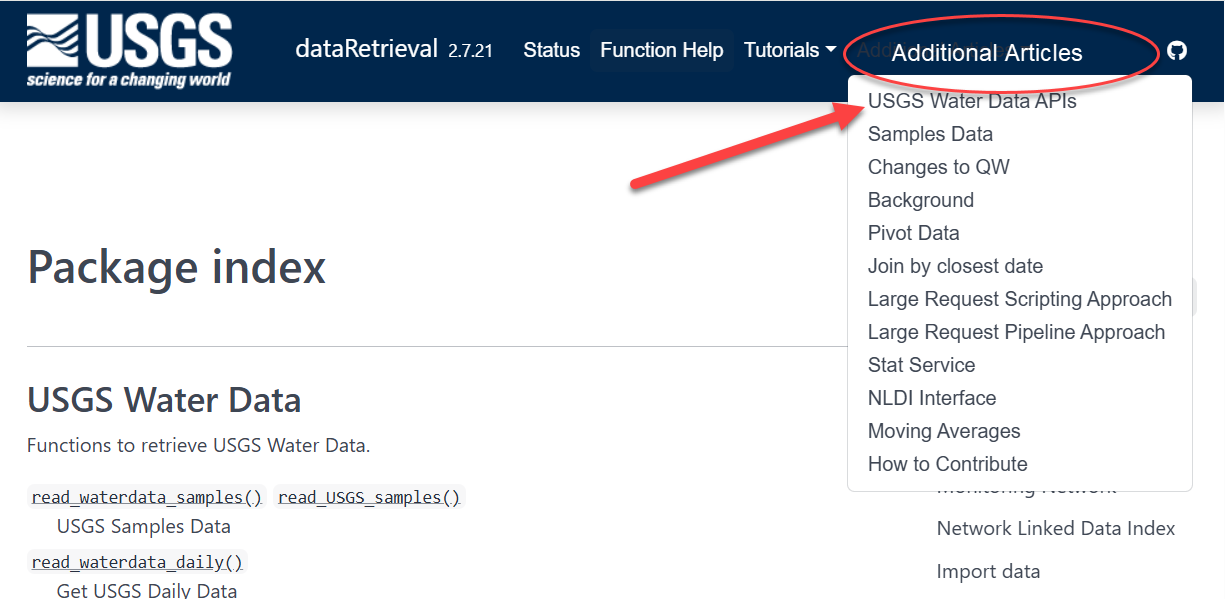
Documentation within R: function help pages
Within R, you can call help files for any dataRetrieval function:
Click here to open a new window:

Exercise 1: Orientation
Open RStudio
Install
dataRetrieval,dplyr,ggplot2, anddata.table(if they are not already installed).Load
dataRetrievalOpen the help file for the function
read_waterdata_dailyNavigate to https://doi-usgs.github.io/dataRetrieval/ and find the list of function help files and explore some articles in “Additional Articles”
dataRetrieval Updates
Are you a seasoned dataRetrieval user?
Here are resources for recent major changes:
What’s New?
There’s been a lot of changes to dataRetrieval over the past year. If you’d like to see an overview of those changes, visit: Changes to dataRetrieval
Biggest changes:
NWIS servers will be shut down, so all
readNWISfunctions will eventually stop workingread_waterdatafunctions are modern and should be used when possibleThe “USGS Water Data APIs” are the new home for USGS data
USGS Water Data API Token
The Water Data APIs limit how many queries a single IP address can make per hour
You can run new
dataRetrievalfunctions without a tokenYou might run into errors quickly. If you (or your IP!) have exceeded the quota, you will see:
! HTTP 429 Too Many Requests.
• You have exceeded your rate limit. Make sure you provided your API key from https://api.waterdata.usgs.gov/signup/, then either try again later or contact us at https://waterdata.usgs.gov/questions-comments/?referrerUrl=https://api.waterdata.usgs.gov for assistance.USGS Water Data API Token
Request a USGS Water Data API Token: https://api.waterdata.usgs.gov/signup/
Save it in a safe place (KeePass or other password management tool)
Add it to your .Renviorn file as API_USGS_PAT.
Restart R
Check that it worked by running (you should see your token printed in the Console):
See next slide for a demonstration.
USGS Water Data API Token: Example
My favorite method to do add your token to .Renviron is to use the usethis package. Let’s pretend the token sent you was “abc123”:
- Run in R:
- Add this line to the file that opens up:
Save that file using the save button
Restart R/RStudio.
Run after restarting R:
USGS Water Data API Token: Example

After save and restart, check that it worked by running:
USGS Basic Retrievals
The USGS uses various codes for basic retrievals. These codes can have leading zeros, therefore they need to be a character surrounded in quotes (“00060”).
- Site ID (often 8 or 15-digits)
- Parameter Code (5 digits)
- Full list:
read_waterdata_parameter_codes()
- Full list:
- Statistic Code (for daily values)
- Full list:
read_metadata("statistic-codes")
- Full list:
USGS Basic Retrievals Parameter and Statistic Codes
Here are some examples of a few common codes:
|
|
Let’s Go!
We’re going walk through 3 retrievals:
Workflow 1: Daily Data
Uses the new USGS Water Data API
Modern data access point going forward
Workflow 2: Discrete Data
Uses new USGS Samples Data
Modern data access point going forward
Workflow 3: Join Daily and Discrete
Workflow 4: Continuous Data
Uses the new USGS Water Data API
Modern data access point going forward
Workflow 5: Join Continuous and Discrete
Workflow 1: Daily data for known site
Let’s pull daily mean discharge data for site “USGS-0940550”, getting all the data from October 10, 2024 onward.
Requesting:
https://api.waterdata.usgs.gov/ogcapi/v0/collections/daily/items?f=json&lang=en-US&monitoring_location_id=USGS-09405500¶meter_code=00060&statistic_id=00003&time=2024-10-01%2F..&limit=50000Remaining requests this hour:2846 Workflow 1: Look at Daily Data
In RStudio, click on the data frame in the upper right Environment tab to open a Viewer.
Workflow 1: Plot Daily Data
Let’s use ggplot2 to visualize the data.
Water Data API Notes: Argument input
Use your “tab” key!
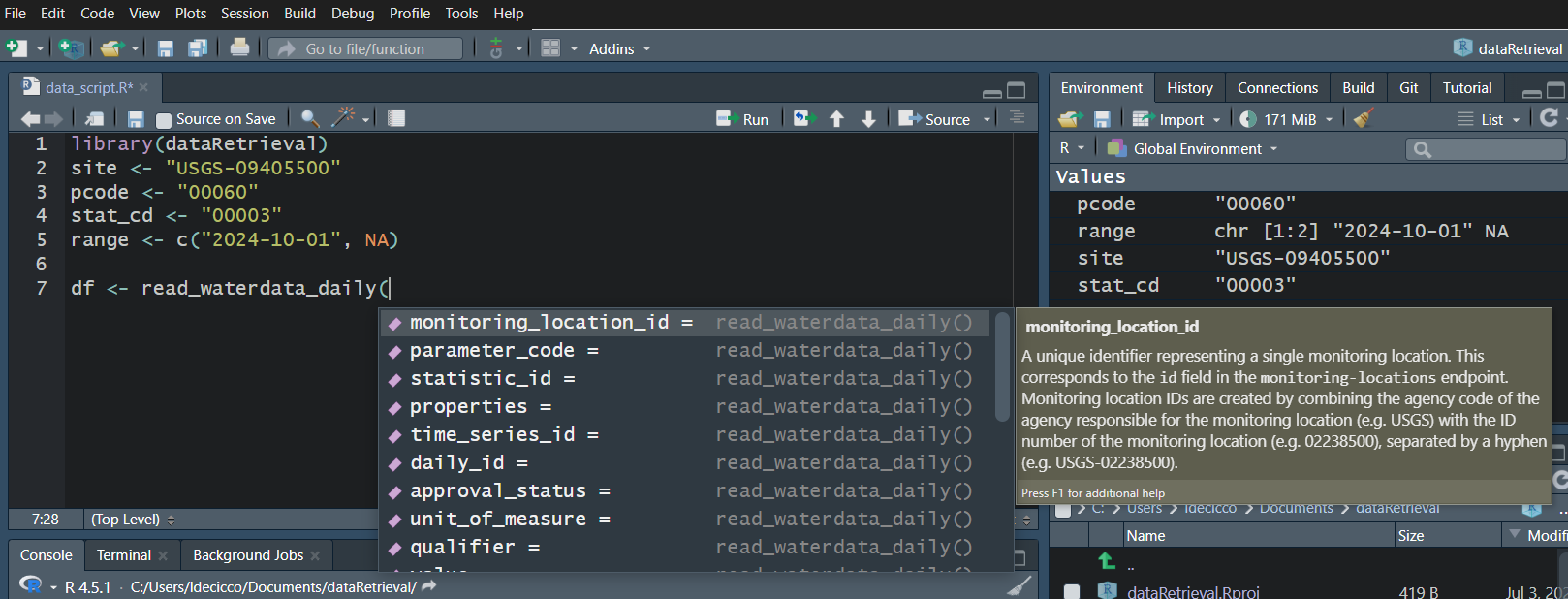
Water Data API Notes: Arguments
When you look at the help file for the new functions, you’ll notice there are lots of possible inputs (arguments).
You DO NOT need to (and should not!) specify all of these parameters.
However, also consider what happens if you leave too many things blank. What do you suppose will be returned here?
Since no list of sites or bounding box was defined, ALL the daily data in ALL the country with parameter code “00060” and statistic code “00003” will be returned.
Water Data API Notes: time input
The “time” argument has a few options:
A single date (or date-time): “2024-10-01” or “2024-10-01T23:20:50Z”
A bounded interval: c(“2024-10-01”, “2025-07-02”)
Half-bounded intervals: c(“2024-10-01”, NA)
Duration objects: “P1M” for data from the past month or “PT36H” for the last 36 hours
Here are a bunch of valid inputs:
# Ask for exact times:
time = "2025-01-01"
time = as.Date("2025-01-01")
time = "2025-01-01T23:20:50Z"
time = as.POSIXct("2025-01-01T23:20:50Z",
format = "%Y-%m-%dT%H:%M:%S",
tz = "UTC")
# Ask for specific range
time = c("2024-01-01", "2025-01-01") # or Dates or POSIXs
# Asking beginning of record to specific end:
time = c(NA, "2024-01-01") # or Date or POSIX
# Asking specific beginning to end of record:
time = c("2024-01-01", NA) # or Date or POSIX
# Ask for period
time = "P1M" # past month
time = "P7D" # past 7 days
time = "PT12H" # past hoursWorkflow 2: Discrete data for known site
Use your “tab” key!
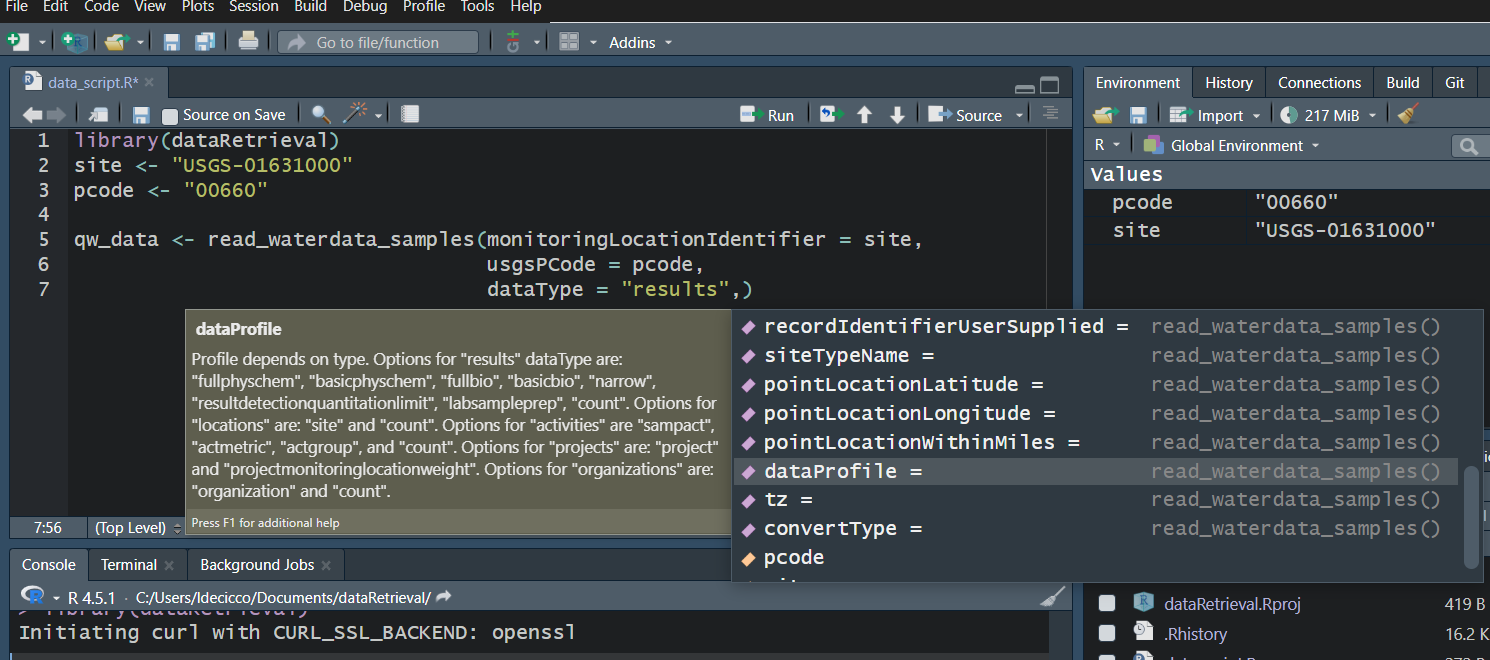
Workflow 2: Discrete data for known site
Let’s get orthophosphate (“00660”) data from the Shenandoah River at Front Royal, VA (“USGS-01631000”).
GET: https://api.waterdata.usgs.gov/samples-data/results/basicphyschem?mimeType=text%2Fcsv&monitoringLocationIdentifier=USGS-01631000&usgsPCode=00660[1] 102That’s a LOT of columns returned. We won’t look at them here, but you can use View in RStudio to explore on your own.
USGS Samples Data Notes: Data Types and Profiles
- There are 2 arguments that dictate what kind of data is returned
- “dataType” defines what kind of data comes back
- “dataProfile” defines what columns from that type come back
Data Types and Profiles
Workflow 2: Discrete data censoring
Let’s pull a few columns out and look at those:
library(dplyr)
qw_data_slim <- qw_data |>
select(Date = Activity_StartDate,
Result_Measure,
DL_cond = Result_ResultDetectionCondition,
DL_val = DetectionLimit_MeasureA,
DL_type = DetectionLimit_TypeA) |>
mutate(Result = if_else(!is.na(DL_cond), DL_val, Result_Measure),
Detected = if_else(!is.na(DL_cond), "Not Detected", "Detected")) |>
arrange(Detected)- What is
|>? It’s a pipe! It says take ‘this thing’ and put it in ‘that thing’. You’ll also see%>%in code, it is also a pipe - they are basically the same.
Workflow 2: Discrete data censoring information
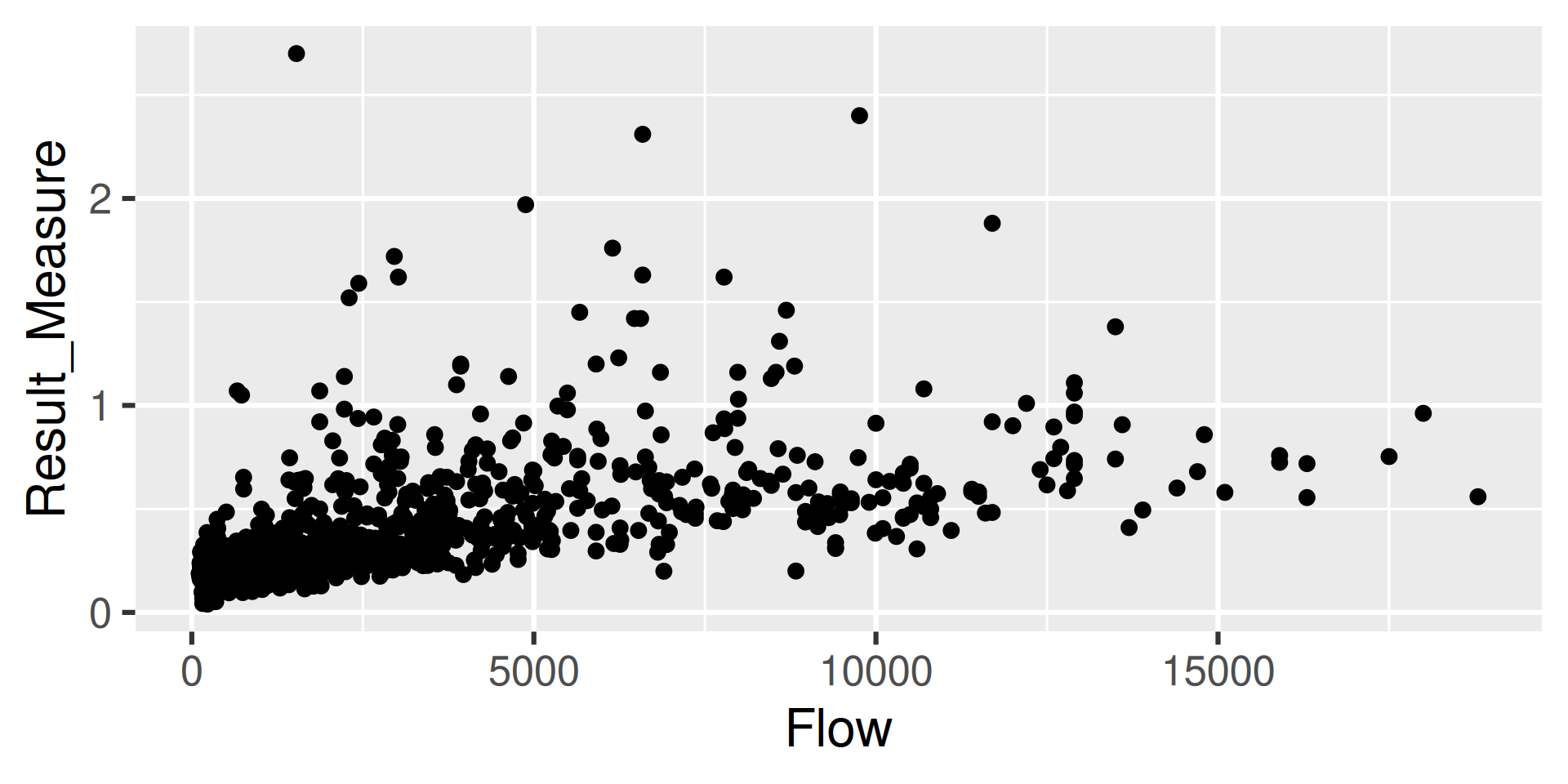
Workflow 3: Join Discrete and Daily
One common workflow is to join discrete data with daily data.
In this example, we will look at a site that measures both water quality parameters and has daily mean discharge.
We will use the
dplyr::left_jointo join the 2 data frames by a date.
Step 1: Get the data
site <- "USGS-04183500"
p_code_dv <- "00060"
stat_cd <- "00003"
p_code_qw <- "00665"
start_date <- "2015-07-03"
end_date <- "2025-07-03"
qw_data <- read_waterdata_samples(monitoringLocationIdentifier = site,
usgsPCode = p_code_qw,
activityStartDateLower = start_date,
activityStartDateUpper = end_date,
dataProfile = "basicphyschem")
dv_data <- read_waterdata_daily(monitoring_location_id = site,
parameter_code = p_code_dv,
statistic_id = stat_cd,
time = c(start_date, end_date))Step 2: Join
- “Activity_StartDate” (on the left side data frame) and “time” (on the right side data frame) need to be the same type (in this case, both are Date objects).
Step 2: Join (cont.)
- You could join on multiple columns:
See dplyr documentation for lots of joining options, but I find left_join my “go-to” for straightforward joins.
Step 3: Inspect
Let’s take a quick peak:
Exercise 2: Joins
dplyr comes with some data sets. To look at them run:
Run that code and view the 2 data frames to see what they look like.
Join the instruments to the “band_members” by name.
Join the members to the “band_instruments” by name.
# A tibble: 3 × 3
name band plays
<chr> <chr> <chr>
1 Mick Stones <NA>
2 John Beatles guitar
3 Paul Beatles bass # A tibble: 3 × 3
name plays band
<chr> <chr> <chr>
1 John guitar Beatles
2 Paul bass Beatles
3 Keith guitar <NA> Workflow 4: Continuous data for known site
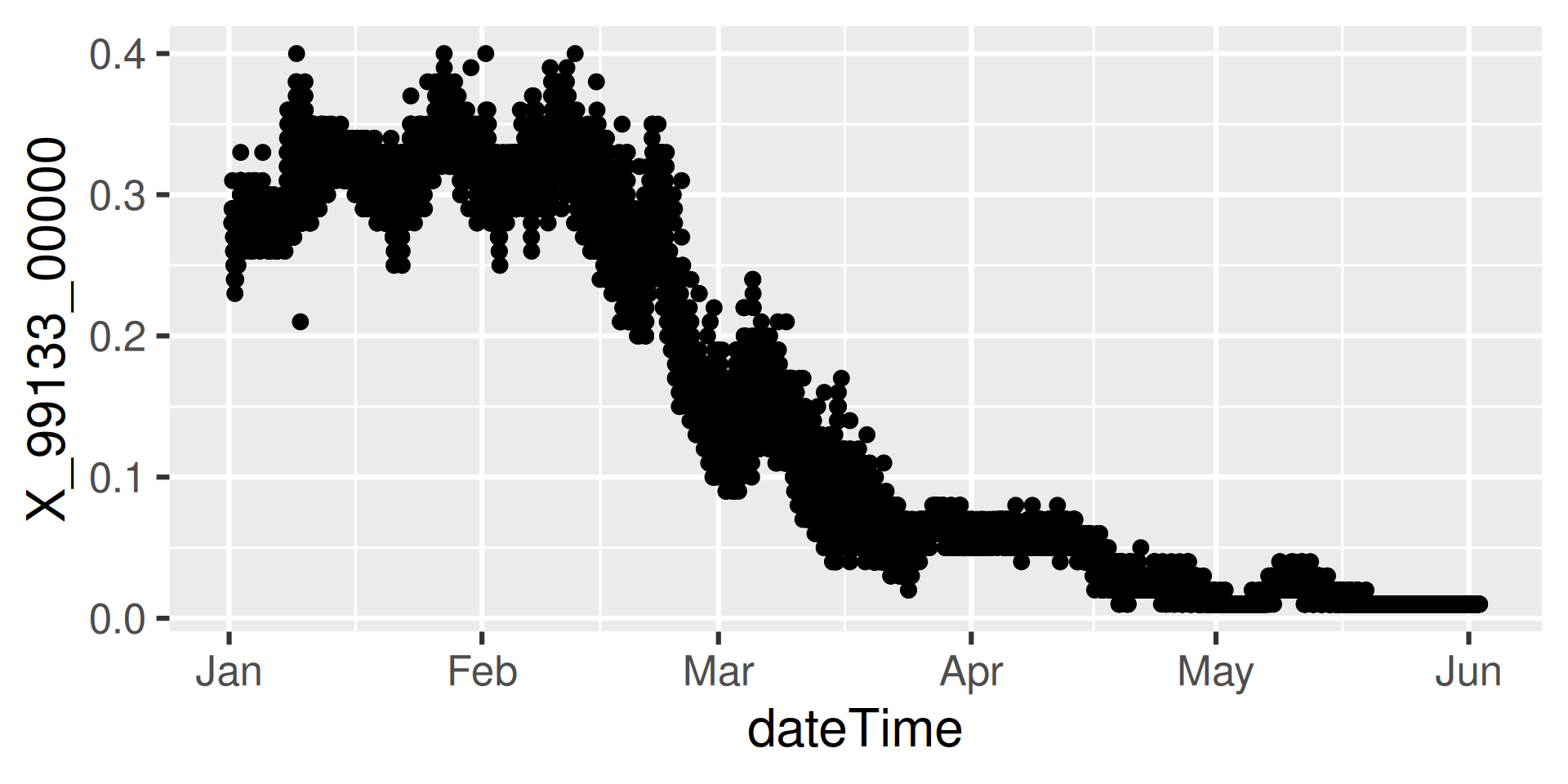
Continuous data is the high-frequency sensor data.
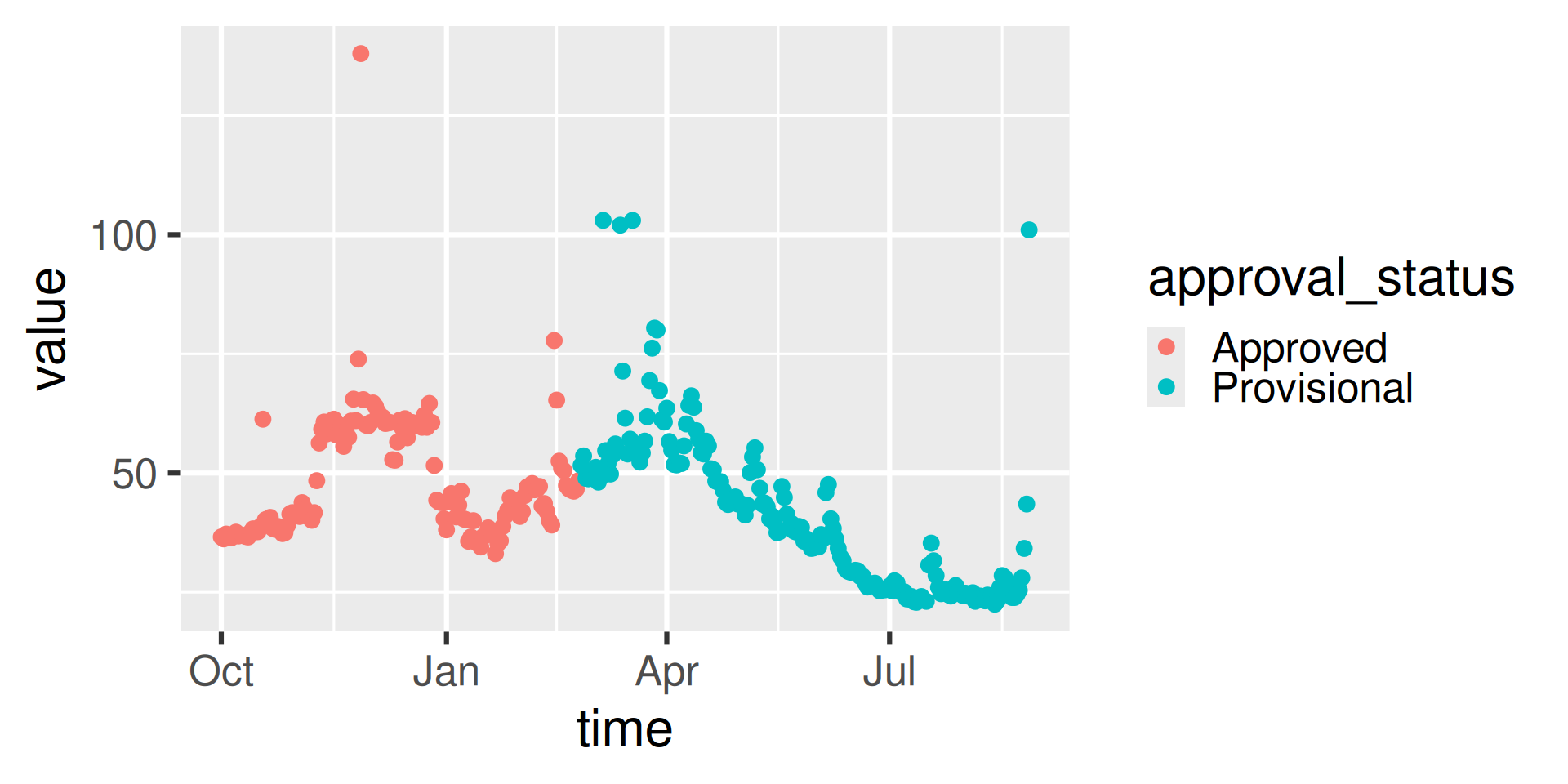
We’ll look at Suisun Bay a Van Sickle Island NR Pittsburg CA (“USGS-11455508”), with parameter code “99133” which is Nitrate plus Nitrite.
Workflow 4: Continuous data for known site
[1] "monitoring_location_id" "parameter_code" "statistic_id"
[4] "time" "value" "unit_of_measure"
[7] "approval_status" "last_modified" "qualifier"
[10] "time_series_id" [4] "time" "unit_of_measure" "parameter_code"
[7] "statistic_id" "value" "approval_status"
[10] "last_modified" "qualifier" Requesting:
https://api.waterdata.usgs.gov/ogcapi/v0/collections/continuous/items?f=json&lang=en-US&skipGeometry=TRUE&limit=50000&monitoring_location_id=USGS-11455508¶meter_code=99133&time=2024-01-01T00%3A00%3A00Z%2F2024-06-01T00%3A00%3A00ZWorkflow 4: Inspect
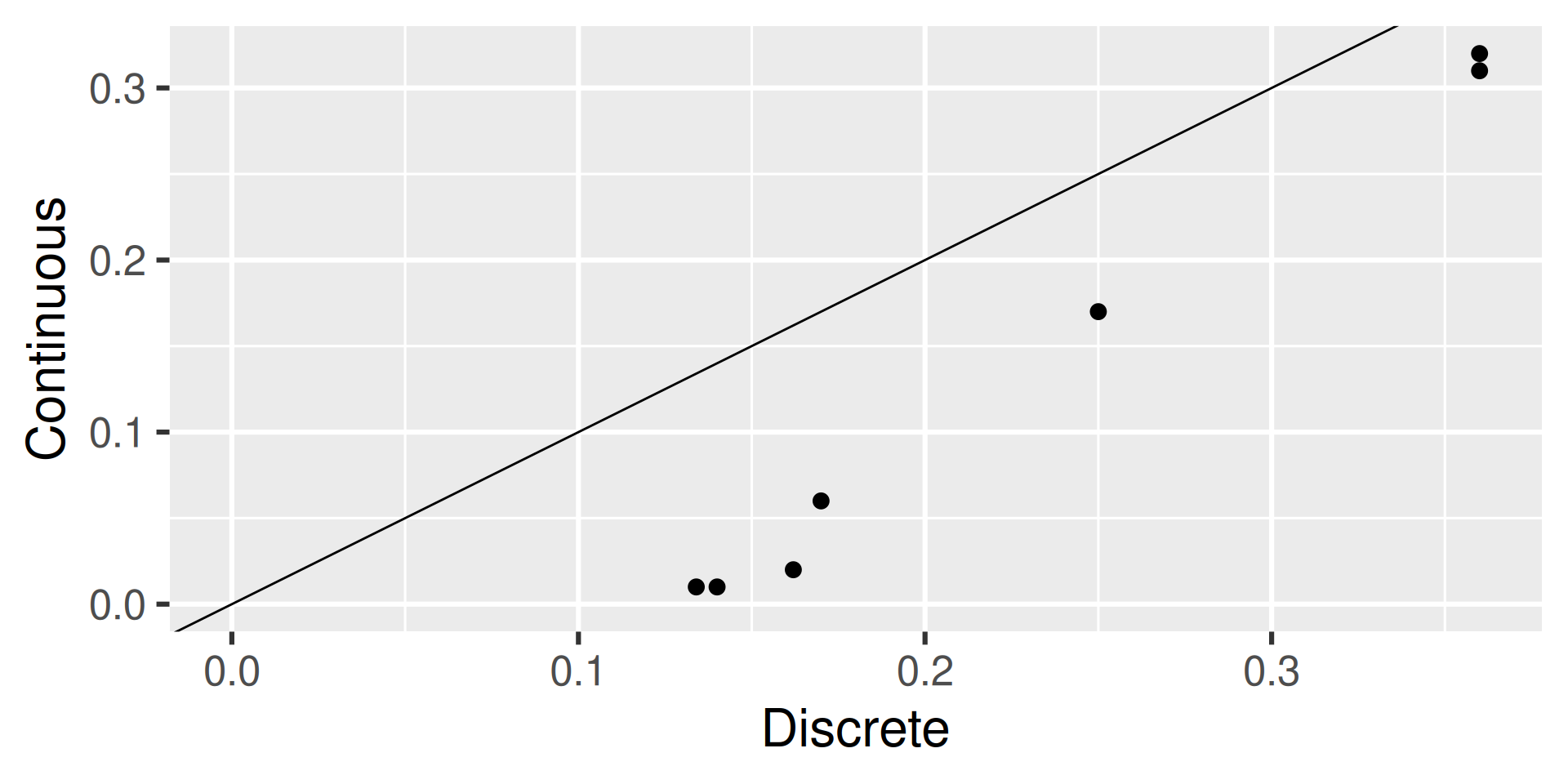
Workflow 5: Join Discrete and Continuous
That same site also measures discrete Nitrate plus Nitrite, which is parameter code “00631”. Let’s first grab that data:
GET: https://api.waterdata.usgs.gov/samples-data/results/basicphyschem?mimeType=text%2Fcsv&monitoringLocationIdentifier=USGS-11455508&usgsPCode=00631&activityStartDateLower=2024-01-01&activityStartDateUpper=2024-06-01Workflow 5: Join Discrete and Continuous
We now want to join the closest continuous sensor time with the discrete sample time.
This is trickier than joining by exact matches.
dplyrhas a way, but it’s complicated if you want the absolute closest in either directionAnother package
data.tablehas a slick way to get the closest matches
Workflow 5: Join Discrete and Continuous
Workflow 5: Inspect
Data Discovery
The process for discovering data is a bit in flux with NWIS retiring. I expect a new process will be introduced soon. For now here are some options.
read_waterdata_ts_metadiscovers daily and continuous time seriessummarize_waterdata_samplesdiscovers discrete data at specific monitoring locations
The next slides will demo how to use those.
Data Discovery: Time Series
Data Discovery: Discrete
characteristicUserSupplied
- characteristicUserSupplied can be an input to
read_waterdata_sample
More Information
- dataRetrieval repository:
- Contact:
- Computational Tools Email: comptools@usgs.gov
Any use of trade, firm, or product name is for descriptive purposes only and does not imply endorsement by the U.S. Government.