Basic usage of SFRmaker in a scripting context
This example illustrates basic usage of SFRmaker in a scripting context. For examples of using SFRmaker with a configuration file, see the MERAS and Tyler Forks examples.
[1]:
import numpy as np
import matplotlib.pyplot as plt
import flopy
import sfrmaker
Input requirements
The most basic input requirements of SFRmaker are hydrography and a model grid. Optionally, a model and a DEM can be input, as demonstrated below. See the documentation for a more detailed description of inputs.
Hydrography
In this example, we will use data that has been downloaded from NHDPlus. The original file structure in the download has been maintained, allowing us to simply supply SFRmaker with a path to the NHDPlus files:
[2]:
NHDPlus_paths = '../tylerforks/NHDPlus/'
If we were dealing with more than one drainage basin, the NHDPlus file paths could be included in a list:
[3]:
NHDPlus_paths_list = ['/NHDPlusGL/NHDPlus04/',
'/NHDPlusMS/NHDPlus07/']
Creating a Lines instance from NHDPlus
The sfrmaker.Lines class includes functionality for reading and processing hydrography flowlines. This example shows how to create a Lines instance from NHDPlus data.
For large hydrography datasets, it is advantageous to filter the data when it is read in. The filter argument to sfrmaker.Lines accepts a shapefile path or tuple of bounding box coordinates. In either case, a bounding box tuple is created and passed to the filter method in the `fiona package <https://fiona.readthedocs.io/en/latest/manual.html>`__, which is fast.
[4]:
lns = sfrmaker.Lines.from_nhdplus_v2(NHDPlus_paths='../tylerforks/NHDPlus/',
bbox_filter='../tylerforks/grid.shp')
loading NHDPlus v2 hydrography data...
for basins:
../tylerforks/NHDPlus/
load finished in 0.11s
Getting routing information from NHDPlus Plusflow table...
finished in 0.02s
Alternatively, sfrmaker.Lines can be instantiated with separate arguments for each NHDPlus file used:
[5]:
lines = sfrmaker.Lines.from_nhdplus_v2(NHDFlowlines='../tylerforks/NHDPlus/NHDSnapshot/Hydrography/NHDFlowline.shp',
PlusFlowlineVAA='../tylerforks/NHDPlus/NHDPlusAttributes/PlusFlowlineVAA.dbf',
PlusFlow='../tylerforks/NHDPlus/NHDPlusAttributes/PlusFlow.dbf',
elevslope='../tylerforks/NHDPlus/NHDPlusAttributes/elevslope.dbf',
bbox_filter=(-90.625, 46.3788, -90.4634, 46.4586))
loading NHDPlus v2 hydrography data...
load finished in 0.04s
Getting routing information from NHDPlus Plusflow table...
finished in 0.02s
Creating a Lines instance from custom hydrography
Alternatively, a Lines instance can be created from any hydrography that includes the pertinent attribute fields, which must be specified:
[6]:
custom_lines = sfrmaker.Lines.from_shapefile(shapefile='../meras/flowlines.shp',
id_column='COMID', # arguments to sfrmaker.Lines.from_shapefile
routing_column='tocomid',
width1_column='width1',
width2_column='width2',
up_elevation_column='elevupsmo',
dn_elevation_column='elevdnsmo',
name_column='GNIS_NAME',
attr_length_units='feet', # units of source data
attr_height_units='feet', # units of source data
)
Specifying a model grid from a flopy StructuredGrid instance
The next step is to specify a model grid. One option is to specify a flopy StructuredGrid instance. SFRmaker will then use this internally to create an instance of its own StructuredGrid class. In this case, the row and column spacing must be provided in the units of the projected coordinate reference system (CRS) that the model is in, which is typically meters. In this case, our model grid spacing is 250 feet, so we have to convert.
Specifying a CRS (via the crs argument) is also important, as it allows SFRmaker to automatically reproject any input data to the same CRS as the model grid. The best way to do this is with an EPSG code, as shown below.
See the flopy documentation for more details about StructuredGrid.
[7]:
delr = np.array([250 * 0.3048] * 160) # cell spacing along a row
delc = np.array([250 * 0.3048] * 111) # cell spacing along a column
flopy_grid = flopy.discretization.StructuredGrid(delr=delr, delc=delc,
xoff=682688, yoff=5139052, # lower left corner of model grid
angrot=0, # grid is unrotated
# projected coordinate system of model (UTM NAD27 zone 15 North)
crs=26715
)
Specifying a model grid from a shapefile
Another option is to create an SFRmaker StructuredGrid directly using a shapefile. While the basic underpinings are in place for SFRmaker to support unstructured grids, this option hasn’t been fully implemented yet.
Attribute fields with row and column information must be specified. An polygon defining the area where the SFR network will be created can optionally be specified here, or later in the creation of SFRData (see below).
[8]:
grid = sfrmaker.StructuredGrid.from_shapefile(shapefile='../tylerforks/grid.shp',
icol='i', # attribute field with row information
jcol='j', # attribute field with column information
active_area='../tylerforks/active_area.shp'
)
reading ../tylerforks/grid.shp...
--> building dataframe... (may take a while for large shapefiles)
reading ../tylerforks/active_area.shp...
--> building dataframe... (may take a while for large shapefiles)
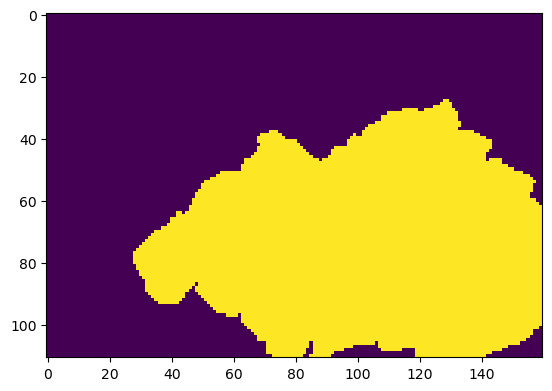
With specification of active_area, the grid created above has an ``isfr`` array attribute designating which cells can have SFR:
[9]:
plt.imshow(grid.isfr, interpolation='nearest')
[9]:
<matplotlib.image.AxesImage at 0x13cb75940>
Using the modelgrid attached to a flopy model
If no grid is supplied as input, SFRmaker will try to use modelgrid attribute attached to a supplied flopy model instance. This only works if modelgrid is valid. Loading a flopy model with a valid model grid requires the grid information to be specified in the namefile header, and that the model be in the same units as the projected CRS. See the flopy documentation for more details
Specifying a model
While a model is not required to run SFRmaker, specifying a model is advantageous in that it allows SFRmaker to assign valid model layers for reaches. Models are specified as flopy model instances, which can be loaded or created from scratch (see the flopy documentation). In this case, we are loading a model:
[10]:
m = flopy.modflow.Modflow.load('tf.nam', model_ws='../tylerforks/tylerforks')
m
[10]:
MODFLOW 5 layer(s) 111 row(s) 160 column(s) 1 stress period(s)
Creating an SFRData instance
The sfrmaker.SFRData class is the primary object for creating or modifying an SFR dataset. A SFRData instance can be created from the Lines class using the to_sfr() method.
Either a flopy StructuredGrid or sfrmaker StructuredGrid can be supplied. While MODFLOW and flopy support specification of length units, these aren’t always specified in the model input files, so it is good practice to specifiy the units explicitly to SFRmaker.
[11]:
sfrdata = lines.to_sfr(grid=flopy_grid, model=m, model_length_units='feet')
SFRmaker version 0.13.0.post8.dev0+gdd6df5f
Creating sfr dataset...
Creating grid class instance from flopy Grid instance...
grid class created in 0.33s
Model grid information
structured grid
nnodes: 17,760
nlay: 1
nrow: 111
ncol: 160
model length units: undefined
crs: EPSG:26715
bounds: 682688.00, 5139052.00, 694880.00, 5147510.20
active area defined by: all cells
MODFLOW 5 layer(s) 111 row(s) 160 column(s) 1 stress period(s)
reprojecting hydrography from
EPSG:4269
to
EPSG:26715
Culling hydrography to active area...
starting lines: 45
remaining lines: 42
finished in 0.00s
Intersecting 42 flowlines with 17,760 grid cells...
Building spatial index...
finished in 0.90s
Intersecting 42 features...
42
finished in 0.10s
Setting up reach data... (may take a few minutes for large grids)
finished in 0.22s
Computing widths...
Dropping 48 reaches with length < 12.50 feet...
Repairing routing connections...
enforcing best segment numbering...
Setting up segment data...
Model grid information
structured grid
nnodes: 17,760
nlay: 1
nrow: 111
ncol: 160
model length units: undefined
crs: EPSG:26715
bounds: 682688.00, 5139052.00, 694880.00, 5147510.20
active area defined by: all cells
Time to create sfr dataset: 1.81s
The SFRData class
Internally, the SFRData class mostly uses the data model for the MODFLOW-2005 style SFR2 package (which includes MODFLOW-NWT), which organizes the input into segments and reaches. Segment and reach data are stored in the reach_data and segment_data attribute DataFrames. In addition to the MODFLOW-2005 data, reach numbers and their routing connections (as needed for MODFLOW-6) are also stored. On writing of SFR package input, MODFLOW-6 style input can be created via the
sfrmaker.mf5to6 module.
[12]:
sfrdata.reach_data.loc[sfrdata.reach_data['rno'] == 1, 'strhc1'] = 5
[13]:
sfrdata.reach_data.head()
[13]:
| rno | node | k | i | j | iseg | ireach | rchlen | width | slope | ... | thts | thti | eps | uhc | outreach | outseg | asum | line_id | name | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 511 | 1 | 17714 | 0 | 110 | 114 | 1 | 1 | 208.807938 | 1.000000 | 0.127779 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 2 | 4 | 31.822330 | 1815013 | None | LINESTRING (691394.6396657807 5139052, 691410.... |
| 512 | 2 | 17715 | 0 | 110 | 115 | 1 | 2 | 294.098938 | 1.261243 | 0.060563 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 3 | 4 | 108.465340 | 1815013 | None | LINESTRING (691451 5139079.0289082145, 691471.... |
| 513 | 3 | 17716 | 0 | 110 | 116 | 1 | 3 | 41.622536 | 1.531957 | 0.333944 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 4 | 4 | 159.629288 | 1815013 | None | LINESTRING (691527.2 5139123.064392628, 691538... |
| 514 | 4 | 17556 | 0 | 109 | 116 | 1 | 4 | 220.366440 | 1.714090 | 0.092205 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 5 | 4 | 199.556412 | 1815013 | None | LINESTRING (691538.8006061709 5139128.2, 69156... |
| 515 | 5 | 17557 | 0 | 109 | 117 | 1 | 5 | 162.617325 | 1.950304 | 0.097331 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 6 | 4 | 257.923126 | 1815013 | None | LINESTRING (691603.4 5139143.66283342, 691624.... |
5 rows × 23 columns
[14]:
sfrdata.segment_data.head()
[14]:
| per | nseg | icalc | outseg | iupseg | iprior | nstrpts | flow | runoff | etsw | ... | uhc1 | hcond2 | thickm2 | elevdn | width2 | depth2 | thts2 | thti2 | eps2 | uhc2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 1 | 4 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1297.867432 | 5.638623 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0 | 2 | 1 | 4 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1297.867432 | 37.791649 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 0 | 3 | 1 | 6 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1278.149658 | 7.069631 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 0 | 4 | 1 | 6 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1278.149658 | 38.638088 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 0 | 5 | 1 | 10 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1211.286133 | 6.470501 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
5 rows × 35 columns
Sampling streambed top elevations from a DEM
While the above dataset contains streambed top elevations read from NHDPlus, a DEM can be sampled to obtain more accurate elevations. If the DEM elevation units are specified, SFRmaker will convert the elevations to model units if needed. See the Streambed_elevation_demo notebook for more information on how this works.
[15]:
sfrdata.set_streambed_top_elevations_from_dem('../tylerforks/dem_26715.tif',
elevation_units='meters')
running rasterstats.zonal_stats on buffered LineStrings...
finished in 4.60s
Smoothing elevations...
finished in 0.11s
Assigning layers to the reaches
Once we have a valid set of streambed elevations, the reaches can be assigned to model layers.
[16]:
sfrdata.assign_layers()
[17]:
sfrdata.reach_data.head()
[17]:
| rno | node | k | i | j | iseg | ireach | rchlen | width | slope | ... | thts | thti | eps | uhc | outreach | outseg | asum | line_id | name | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 511 | 1 | 17714 | 2 | 110 | 114 | 1 | 1 | 208.807938 | 1.000000 | 0.034189 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 2 | 4 | 31.822330 | 1815013 | None | LINESTRING (691394.6396657807 5139052, 691410.... |
| 512 | 2 | 17715 | 2 | 110 | 115 | 1 | 2 | 294.098938 | 1.261243 | 0.000750 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 3 | 4 | 108.465340 | 1815013 | None | LINESTRING (691451 5139079.0289082145, 691471.... |
| 513 | 3 | 17716 | 1 | 110 | 116 | 1 | 3 | 41.622536 | 1.531957 | 0.165061 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 4 | 4 | 159.629288 | 1815013 | None | LINESTRING (691527.2 5139123.064392628, 691538... |
| 514 | 4 | 17556 | 2 | 109 | 116 | 1 | 4 | 220.366440 | 1.714090 | 0.027078 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 5 | 4 | 199.556412 | 1815013 | None | LINESTRING (691538.8006061709 5139128.2, 69156... |
| 515 | 5 | 17557 | 2 | 109 | 117 | 1 | 5 | 162.617325 | 1.950304 | 0.023848 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 6 | 4 | 257.923126 | 1815013 | None | LINESTRING (691603.4 5139143.66283342, 691624.... |
5 rows × 23 columns
Running diagnostics
SFRData includes a run_diagnostics() method that executes the Flopy checker on the SFR package input. While the Flopy checks are only implemented for MODFLOW-2005 style SFR packages, run_diagnostics() works on MODFLOW-6 packages as well, by working of an attached MODFLOW-2005 representation of the MODFLOW-6 SFR input. The following checks are executed:
NaNs (not a number values, which will cause MODFLOW to crash)
consecutive segment and reach numbering
segment numbering that only increases in the downstream direction
circular routing sequences (reaches or segments routing back to themselves)
routing connection proximity: The length of routing connections, as determined by the distance between their cell centers, is compared to 1.25 times the diagonal distance across the cell containing the upstream reach. This benchmark length is equivalent to a diagonal connection between two square cells, where one cell had a spacing of 1.5 times the other cell (the maximum recommended increase in spacing between two adjacent cells; see Anderson and others, 2015). While this routing proximity check can be helpful for identifying egregious routing issues, in practice, it may not be uncommon for valid routing connections to violate this rule. For example, if the
one_reach_per_celloption is used in constructing the SFR package with a large grid size, there may be numerous routing connections that extend across several cells. When non-adjacent routing connections are identified, the user can check the shapefile of routing connections to verify the results.overlapping conductances: This check identifies instances of collocated reaches, where more than one reach has a connection to the groundwater flow solution in that cell. Collocated reaches may or may not present an issue for a particular model solution, but can potentially promote instability or spurious circulation of flow between the collocated reaches.
spurious streambed top elevations: Values < -10 or > 15,000 are flagged
downstream rises in streambed elevation
inconsistencies between streambed elevation and the model grid: This check looks for streambed bottoms below their respective cell bottoms, which will cause MODFLOW to halt execution, and streambed tops that are above the model top. The latter issue will not prevent MODFLOW from running, but may indicate problems with the input if the differences are large. For coarse grids in areas of high stream density (where there are often multiple streams in a cell), it may be impossible to ensure that all streambed top elevations are at or below the model top. Oftentimes, these reaches are dry anyways (for example, and ephemeral gully that runs off a bluff into a perennial stream, where the cell elevation is based on the elevation of the perennial stream). In any case, it is good practice to check the largest violations by comparing the SFRmaker shapefile output to a DEM and other data in a GIS environment.
suprious slopes: slopes of less than 0.0001 (which can lead to excessive stages with the SFR package’s Manning approximation) and greater than 1.0 are flagged.
[18]:
sfrdata.run_diagnostics(verbose=False)
Running Flopy v. 3.10.0 diagnostics...
wrote tf_SFR.chk
Writing an SFR package
Now we can write the package. By default, a MODFLOW-2005 style package is written to the current working directory or the model workspace of the attached model instance.
[19]:
sfrdata.write_package()
SFRmaker v. 0.13.0.post8.dev0+gdd6df5f
Running Flopy v. 3.10.0 diagnostics...
passed.
Checking for continuity in segment and reach numbering...
passed.
Checking for increasing segment numbers in downstream direction...
passed.
Checking for circular routing...
passed.
Checking reach connections for proximity...
5 segments with non-adjacent reaches found.
At segments:
41 20 21 22 23
5 segments with non-adjacent reaches found.
At segments:
41 20 21 22 23
Checking for model cells with multiple non-zero SFR conductances...
24 model cells with multiple non-zero SFR conductances found.
This may lead to circular routing between collocated reaches.
Nodes with overlapping conductances:
k i j iseg ireach rchlen strthick strhc1
2 110 119 1 8 268.5836486816406 1.0 1.0
1 110 119 2 1 72.61438751220703 1.0 1.0
0 87 122 3 54 35.35249328613281 1.0 1.0
0 87 122 4 33 231.09567260742188 1.0 1.0
0 76 110 5 47 204.88766479492188 1.0 1.0
0 87 122 6 1 30.414600372314453 1.0 1.0
2 84 119 6 7 271.24835205078125 1.0 1.0
2 84 119 6 9 26.37481117248535 1.0 1.0
0 76 110 6 27 75.33216094970703 1.0 1.0
2 25 24 7 42 95.27275085449219 1.0 1.0
2 25 24 8 47 230.57635498046875 1.0 1.0
1 58 132 9 25 48.14010238647461 1.0 1.0
1 58 132 9 27 92.92947387695312 1.0 1.0
0 48 109 9 64 251.07447814941406 1.0 1.0
0 76 110 10 1 69.20076751708984 1.0 1.0
0 48 108 10 40 19.56378746032715 1.0 1.0
0 48 109 10 41 122.84815216064453 1.0 1.0
0 48 108 10 42 28.33331871032715 1.0 1.0
2 25 24 11 1 21.51937484741211 1.0 1.0
2 18 17 11 15 148.96444702148438 1.0 1.0
2 18 17 12 57 76.79739379882812 1.0 1.0
1 80 49 13 70 159.26600646972656 1.0 1.0
0 48 108 14 1 125.24523162841797 1.0 1.0
0 47 98 14 22 67.60189056396484 1.0 1.0
0 47 98 14 24 103.03018188476562 1.0 1.0
0 54 89 14 39 34.45417785644531 1.0 1.0
0 54 89 14 41 60.66257095336914 1.0 1.0
0 53 78 14 57 61.411861419677734 1.0 1.0
0 53 78 14 59 28.438196182250977 1.0 1.0
0 52 76 14 62 63.360740661621094 1.0 1.0
0 52 76 14 64 294.8589782714844 1.0 1.0
1 54 73 14 69 341.6566162109375 1.0 1.0
1 54 73 14 71 27.671581268310547 1.0 1.0
1 80 49 14 125 39.762516021728516 1.0 1.0
2 18 17 16 1 185.54652404785156 1.0 1.0
2 15 3 16 21 32.64251708984375 1.0 1.0
1 36 11 17 38 57.836700439453125 1.0 1.0
2 110 159 19 2 201.63583374023438 1.0 1.0
0 38 85 21 8 72.7103042602539 1.0 1.0
0 38 85 21 9 54.39541244506836 1.0 1.0
2 108 57 24 8 237.04415893554688 1.0 1.0
2 108 57 24 9 46.142940521240234 1.0 1.0
0 94 35 24 45 96.94855499267578 1.0 1.0
0 94 35 24 47 91.54985046386719 1.0 1.0
2 91 35 24 50 60.89990997314453 1.0 1.0
2 91 34 24 51 61.16813278198242 1.0 1.0
1 80 49 25 1 146.93106079101562 1.0 1.0
2 91 35 25 26 308.2864685058594 1.0 1.0
2 91 34 25 27 75.82389068603516 1.0 1.0
2 15 3 31 1 273.603271484375 1.0 1.0
2 36 11 36 1 263.1260681152344 1.0 1.0
2 110 159 38 1 24.2677001953125 1.0 1.0
2 91 34 42 1 130.52822875976562 1.0 1.0
1 92 30 42 6 66.65867614746094 1.0 1.0
1 92 30 42 8 252.91909790039062 1.0 1.0
Checking for streambed tops of less than -10...
passed.
Checking for streambed tops of greater than 15000...
passed.
Checking segment_data for downstream rises in streambed elevation...
Segment elevup and elevdn not specified for nstrm=-1439 and isfropt=1
passed.
Checking reach_data for downstream rises in streambed elevation...
passed.
Checking reach_data for inconsistencies between streambed elevations and the model grid...
186 reaches encountered with streambed above model top.
Model top violations:
i j iseg ireach strtop modeltop strhc1 reachID diff
44 51 8 2 1096.0574951171875 1084.9100341796875 1.0 214 -11.1474609375
31 53 23 10 1008.349853515625 998.0900268554688 1.0 931 -10.25982666015625
34 50 12 5 1016.0361328125 1006.780029296875 1.0 385 -9.256103515625
42 49 8 6 1066.8226318359375 1057.7900390625 1.0 218 -9.0325927734375
3 76 27 3 1009.2495727539062 1002.1099853515625 1.0 1082 -7.13958740234375
94 3 37 21 1073.4644775390625 1066.4599609375 1.0 1360 -7.0045166015625
61 22 18 2 1029.281982421875 1022.3200073242188 1.0 769 -6.96197509765625
84 111 5 34 1261.253662109375 1254.3699951171875 1.0 130 -6.8836669921875
47 29 15 21 965.2784423828125 958.4199829101562 1.0 653 -6.85845947265625
61 18 33 3 996.3890991210938 989.6300048828125 1.0 1246 -6.75909423828125
71 5 35 9 990.4193725585938 983.8200073242188 1.0 1310 -6.599365234375
43 50 8 4 1078.9158935546875 1072.3599853515625 1.0 216 -6.555908203125
84 9 34 2 1085.3753662109375 1079.010009765625 1.0 1287 -6.3653564453125
35 51 12 3 1028.656494140625 1022.8200073242188 1.0 383 -5.83648681640625
108 54 24 12 1228.1346435546875 1222.3299560546875 1.0 1010 -5.8046875
33 4 36 10 834.1530151367188 828.52001953125 1.0 1334 -5.63299560546875
0 100 40 1 1027.534423828125 1022.1199951171875 1.0 1384 -5.4144287109375
5 25 28 45 817.1793823242188 811.8099975585938 1.0 1135 -5.369384765625
22 12 15 62 824.2842407226562 819.0800170898438 1.0 694 -5.2042236328125
52 37 15 9 1054.0181884765625 1048.8399658203125 1.0 641 -5.17822265625
65 1 35 19 944.7957153320312 939.6599731445312 1.0 1320 -5.1357421875
49 15 18 19 925.3494262695312 920.3099975585938 1.0 786 -5.0394287109375
51 24 17 11 963.3839721679688 958.3800048828125 1.0 740 -5.00396728515625
26 47 23 21 942.5027465820312 937.5900268554688 1.0 942 -4.9127197265625
1 23 28 51 778.961669921875 774.219970703125 1.0 1141 -4.74169921875
73 5 35 7 1003.2875366210938 998.5999755859375 1.0 1308 -4.68756103515625
45 28 15 24 947.1143188476562 942.72998046875 1.0 656 -4.38433837890625
80 116 6 18 1229.7757568359375 1225.4599609375 1.0 161 -4.3157958984375
27 18 15 51 863.89599609375 859.6300048828125 1.0 683 -4.2659912109375
82 112 5 39 1242.8052978515625 1238.5400390625 1.0 135 -4.2652587890625
35 31 7 25 922.0911254882812 918.2000122070312 1.0 195 -3.89111328125
30 52 23 12 991.4169921875 987.5700073242188 1.0 933 -3.84698486328125
71 4 35 10 981.303466796875 977.6799926757812 1.0 1311 -3.62347412109375
58 19 18 6 986.8626708984375 983.260009765625 1.0 773 -3.6026611328125
87 110 5 30 1283.4881591796875 1279.989990234375 1.0 126 -3.4981689453125
59 17 33 5 976.4671630859375 973.0399780273438 1.0 1248 -3.42718505859375
85 111 5 33 1264.7679443359375 1261.3499755859375 1.0 129 -3.41796875
41 12 18 31 892.8302001953125 889.469970703125 1.0 798 -3.3602294921875
54 12 33 15 932.0604248046875 928.719970703125 1.0 1258 -3.3404541015625
16 32 23 46 867.0645751953125 863.75 1.0 967 -3.3145751953125
28 40 12 21 934.5240478515625 931.27001953125 1.0 401 -3.2540283203125
1 138 26 1 1154.71630859375 1151.489990234375 1.0 1077 -3.226318359375
10 25 23 59 824.5831909179688 821.3699951171875 1.0 980 -3.21319580078125
36 32 7 23 927.737060546875 924.5399780273438 1.0 193 -3.19708251953125
51 15 18 17 930.7290649414062 927.5700073242188 1.0 784 -3.1590576171875
67 2 35 16 953.3693237304688 950.25 1.0 1317 -3.11932373046875
82 140 3 27 1305.33447265625 1302.239990234375 1.0 36 -3.094482421875
58 4 32 22 912.39599609375 909.3099975585938 1.0 1234 -3.08599853515625
31 28 7 32 902.4729614257812 899.4500122070312 1.0 202 -3.02294921875
18 0 30 40 762.85107421875 759.8400268554688 1.0 1204 -3.01104736328125
88 110 5 29 1288.3612060546875 1285.3800048828125 1.0 125 -2.981201171875
34 30 7 27 917.8261108398438 914.8599853515625 1.0 197 -2.96612548828125
33 13 30 13 879.4591674804688 876.5599975585938 1.0 1177 -2.899169921875
1 137 26 2 1149.9388427734375 1147.0799560546875 1.0 1078 -2.85888671875
109 58 24 6 1244.657470703125 1241.8199462890625 1.0 1004 -2.8375244140625
6 22 23 66 783.7332153320312 780.9299926757812 1.0 987 -2.80322265625
28 7 30 24 838.6741943359375 835.8900146484375 1.0 1188 -2.7841796875
83 51 13 65 1143.177490234375 1140.4300537109375 1.0 502 -2.7474365234375
20 9 15 67 797.5311279296875 794.7999877929688 1.0 699 -2.73114013671875
83 145 3 21 1313.221923828125 1310.52001953125 1.0 30 -2.701904296875
34 43 8 20 971.7984619140625 969.0999755859375 1.0 232 -2.698486328125
43 39 7 10 1002.135009765625 999.4600219726562 1.0 180 -2.67498779296875
18 16 16 2 816.8546142578125 814.1799926757812 1.0 710 -2.67462158203125
80 2 34 13 1005.6331176757812 1002.989990234375 1.0 1298 -2.64312744140625
57 27 17 2 1030.468994140625 1027.8399658203125 1.0 731 -2.6290283203125
98 6 37 14 1130.2991943359375 1127.739990234375 1.0 1353 -2.5592041015625
96 6 37 16 1109.81494140625 1107.27001953125 1.0 1355 -2.544921875
41 27 15 29 922.3822631835938 919.8499755859375 1.0 661 -2.53228759765625
93 35 24 48 1133.5345458984375 1131.030029296875 1.0 1046 -2.5045166015625
11 26 23 57 835.0635986328125 832.5999755859375 1.0 978 -2.463623046875
57 18 18 8 976.854248046875 974.4600219726562 1.0 775 -2.39422607421875
43 27 15 27 935.5574951171875 933.1699829101562 1.0 659 -2.38751220703125
29 19 15 48 875.1234130859375 872.77001953125 1.0 680 -2.3533935546875
97 6 37 15 1122.60205078125 1120.280029296875 1.0 1354 -2.322021484375
76 110 6 27 1202.379638671875 1200.0799560546875 1.0 170 -2.2996826171875
13 29 23 52 853.8441162109375 851.5800170898438 1.0 973 -2.26409912109375
28 50 23 16 969.1011352539062 966.8800048828125 1.0 937 -2.22113037109375
41 0 33 40 858.0049438476562 855.7899780273438 1.0 1283 -2.2149658203125
25 16 15 55 848.8750610351562 846.7100219726562 1.0 687 -2.1650390625
3 24 28 48 797.5549926757812 795.4099731445312 1.0 1138 -2.14501953125
65 10 32 9 973.0398559570312 970.9299926757812 1.0 1221 -2.10986328125
98 100 5 9 1381.1871337890625 1379.1099853515625 1.0 105 -2.0771484375
21 25 12 43 869.5907592773438 867.52001953125 1.0 423 -2.07073974609375
78 111 5 44 1220.5587158203125 1218.530029296875 1.0 140 -2.0286865234375
55 25 17 6 996.66845703125 994.6400146484375 1.0 735 -2.0284423828125
51 36 15 11 1035.9466552734375 1033.9599609375 1.0 643 -1.9866943359375
30 34 8 32 918.734375 916.7899780273438 1.0 244 -1.94439697265625
32 21 15 44 887.8782348632812 885.9400024414062 1.0 676 -1.938232421875
4 77 27 1 1019.9188232421875 1018.0 1.0 1080 -1.9188232421875
51 9 33 21 916.0432739257812 914.1300048828125 1.0 1264 -1.91326904296875
27 6 30 26 830.7896118164062 828.9600219726562 1.0 1190 -1.82958984375
6 26 28 43 833.5056762695312 831.719970703125 1.0 1133 -1.78570556640625
22 2 30 34 786.0257568359375 784.25 1.0 1198 -1.7757568359375
7 26 28 42 844.1419677734375 842.3800048828125 1.0 1132 -1.761962890625
48 6 33 27 898.0303955078125 896.2899780273438 1.0 1270 -1.74041748046875
18 21 12 51 847.4663696289062 845.739990234375 1.0 431 -1.72637939453125
89 64 13 41 1181.4326171875 1179.72998046875 1.0 478 -1.70263671875
74 6 35 5 1011.9109497070312 1010.219970703125 1.0 1306 -1.69097900390625
8 69 22 4 1005.525390625 1003.8599853515625 1.0 904 -1.6654052734375
13 45 28 18 909.0931396484375 907.5 1.0 1108 -1.5931396484375
43 13 18 28 900.6517333984375 899.0599975585938 1.0 795 -1.59173583984375
46 14 18 24 910.212890625 908.7100219726562 1.0 791 -1.50286865234375
15 6 16 16 769.787109375 768.2899780273438 1.0 724 -1.49713134765625
3 39 29 12 893.4083862304688 891.9400024414062 1.0 1155 -1.4683837890625
84 145 3 20 1316.32958984375 1314.9100341796875 1.0 29 -1.4195556640625
12 28 23 54 846.7885131835938 845.3699951171875 1.0 975 -1.41851806640625
20 1 30 37 775.9326171875 774.530029296875 1.0 1201 -1.402587890625
47 5 33 29 893.9849243164062 892.5900268554688 1.0 1272 -1.3948974609375
57 3 32 24 906.1943359375 904.8900146484375 1.0 1236 -1.3043212890625
37 20 30 2 917.4218139648438 916.1199951171875 1.0 1166 -1.30181884765625
76 110 5 47 1201.3443603515625 1200.0799560546875 1.0 143 -1.264404296875
76 110 10 1 1201.3443603515625 1200.0799560546875 1.0 324 -1.264404296875
68 3 35 14 962.5520629882812 961.3599853515625 1.0 1315 -1.19207763671875
51 119 9 45 1191.214111328125 1190.050048828125 1.0 304 -1.1640625
25 46 23 23 935.9991455078125 934.8499755859375 1.0 944 -1.149169921875
39 26 15 32 915.0709838867188 913.97998046875 1.0 664 -1.09100341796875
70 3 35 12 969.0398559570312 967.9500122070312 1.0 1313 -1.08984375
6 158 39 3 1123.38037109375 1122.300048828125 1.0 1372 -1.080322265625
37 25 15 35 908.1263427734375 907.0700073242188 1.0 667 -1.05633544921875
26 25 7 40 879.2858276367188 878.239990234375 1.0 210 -1.04583740234375
97 41 24 35 1150.635498046875 1149.6500244140625 1.0 1033 -0.9854736328125
60 107 10 26 1175.4578857421875 1174.52001953125 1.0 349 -0.9378662109375
46 21 17 19 923.8571166992188 922.9199829101562 1.0 748 -0.9371337890625
25 158 20 3 1172.1849365234375 1171.27001953125 1.0 807 -0.9149169921875
39 48 8 10 1029.3406982421875 1028.449951171875 1.0 222 -0.8907470703125
14 99 21 61 1097.334716796875 1096.469970703125 1.0 879 -0.86474609375
40 86 21 5 1143.69384765625 1142.8499755859375 1.0 823 -0.8438720703125
85 52 13 62 1149.013427734375 1148.1700439453125 1.0 499 -0.8433837890625
52 109 10 35 1164.787353515625 1163.969970703125 1.0 358 -0.8173828125
38 12 17 36 880.8272094726562 880.030029296875 1.0 765 -0.79718017578125
74 8 35 3 1025.747314453125 1024.9599609375 1.0 1304 -0.787353515625
105 51 24 18 1209.4859619140625 1208.699951171875 1.0 1016 -0.7860107421875
51 108 10 37 1162.79296875 1162.010009765625 1.0 360 -0.782958984375
25 33 12 31 903.5383911132812 902.760009765625 1.0 411 -0.77838134765625
28 25 7 38 886.2448120117188 885.5 1.0 208 -0.74481201171875
0 71 27 11 967.5951538085938 966.8599853515625 1.0 1090 -0.73516845703125
66 10 32 8 977.9095458984375 977.219970703125 1.0 1220 -0.6895751953125
5 157 39 5 1121.365966796875 1120.6800537109375 1.0 1374 -0.6859130859375
45 13 18 26 906.5974731445312 905.9299926757812 1.0 793 -0.66748046875
26 158 20 2 1174.694580078125 1174.030029296875 1.0 806 -0.66455078125
62 19 33 1 1005.5029296875 1004.8400268554688 1.0 1244 -0.66290283203125
56 18 18 9 967.2000732421875 966.5399780273438 1.0 776 -0.66009521484375
33 80 21 19 1128.41845703125 1127.780029296875 1.0 837 -0.638427734375
101 41 24 31 1158.5797119140625 1157.969970703125 1.0 1029 -0.6097412109375
74 109 10 4 1198.0435791015625 1197.43994140625 1.0 327 -0.6036376953125
30 10 30 19 857.75048828125 857.1599731445312 1.0 1183 -0.59051513671875
96 36 24 42 1141.318359375 1140.72998046875 1.0 1040 -0.58837890625
33 22 15 42 892.9962768554688 892.4299926757812 1.0 674 -0.5662841796875
6 104 21 74 1082.59326171875 1082.06005859375 1.0 892 -0.533203125
104 44 24 26 1173.2772216796875 1172.8199462890625 1.0 1024 -0.457275390625
42 27 15 28 927.3306884765625 926.8800048828125 1.0 660 -0.45068359375
26 17 15 53 855.5484008789062 855.1099853515625 1.0 685 -0.43841552734375
93 122 4 27 1282.7025146484375 1282.2900390625 1.0 90 -0.4124755859375
20 89 21 45 1108.5118408203125 1108.0999755859375 1.0 863 -0.411865234375
32 29 7 30 907.5672607421875 907.1599731445312 1.0 200 -0.40728759765625
6 44 29 4 923.0087890625 922.6599731445312 1.0 1147 -0.34881591796875
98 41 24 34 1152.4619140625 1152.1199951171875 1.0 1032 -0.3419189453125
60 6 32 18 933.5938720703125 933.260009765625 1.0 1230 -0.3338623046875
37 21 30 1 919.2725830078125 918.9400024414062 1.0 1165 -0.33258056640625
89 122 4 31 1278.8204345703125 1278.5 1.0 94 -0.3204345703125
57 108 10 30 1171.8583984375 1171.5400390625 1.0 353 -0.318359375
34 81 21 17 1129.405517578125 1129.0899658203125 1.0 835 -0.3155517578125
82 50 13 67 1139.371337890625 1139.06005859375 1.0 504 -0.311279296875
53 108 10 34 1166.6656494140625 1166.4000244140625 1.0 357 -0.265625
88 55 13 55 1161.44287109375 1161.199951171875 1.0 492 -0.242919921875
47 105 14 7 1156.3297119140625 1156.0999755859375 1.0 514 -0.229736328125
58 108 10 29 1172.513916015625 1172.2900390625 1.0 352 -0.223876953125
99 123 4 18 1290.2703857421875 1290.0999755859375 1.0 81 -0.17041015625
101 95 5 1 1400.4940185546875 1400.3399658203125 1.0 97 -0.154052734375
36 19 30 4 910.9712524414062 910.8200073242188 1.0 1168 -0.1512451171875
86 52 13 61 1151.74169921875 1151.5999755859375 1.0 498 -0.1417236328125
23 2 30 33 790.0592651367188 789.9199829101562 1.0 1197 -0.1392822265625
36 46 8 15 997.1134033203125 996.97998046875 1.0 227 -0.1334228515625
15 31 23 48 859.490234375 859.3599853515625 1.0 969 -0.1302490234375
16 3 15 76 757.1549682617188 757.0399780273438 1.0 708 -0.114990234375
99 14 37 5 1152.94384765625 1152.8499755859375 1.0 1344 -0.0938720703125
89 56 13 53 1164.082763671875 1163.989990234375 1.0 490 -0.0927734375
91 122 4 29 1280.729736328125 1280.6400146484375 1.0 92 -0.0897216796875
35 58 23 1 1069.680419921875 1069.5999755859375 1.0 922 -0.0804443359375
52 81 14 51 1142.7772216796875 1142.699951171875 1.0 558 -0.0772705078125
46 120 9 51 1181.7955322265625 1181.719970703125 1.0 310 -0.0755615234375
40 48 8 9 1038.634765625 1038.56005859375 1.0 221 -0.07470703125
99 41 24 33 1153.8238525390625 1153.75 1.0 1031 -0.0738525390625
75 110 10 2 1199.30908203125 1199.25 1.0 325 -0.05908203125
47 120 9 50 1182.4256591796875 1182.3699951171875 1.0 309 -0.0556640625
98 123 4 19 1289.3104248046875 1289.2900390625 1.0 82 -0.0203857421875
Checking segment_data for inconsistencies between segment end elevations and the model grid...
Segment elevup and elevdn not specified for nstrm=-1439 and isfropt=1
passed.
Checking for streambed slopes of less than 0.0001...
passed.
Checking for streambed slopes of greater than 1.0...
passed.
wrote tf_SFR.chk
wrote /Users/runner/work/sfrmaker/sfrmaker/examples/Notebooks/../tylerforks/tylerforks/tf.sfr.
Writing a MODFLOW-6 SFR package
Alternatively, a MODFLOW-6 style SFR package can be written by specifying version='mf6'
[20]:
sfrdata.write_package('tf_mf6.sfr', version='mf6')
SFRmaker v. 0.13.0.post8.dev0+gdd6df5f
Running Flopy v. 3.10.0 diagnostics...
passed.
Checking for continuity in segment and reach numbering...
passed.
Checking for increasing segment numbers in downstream direction...
passed.
Checking for circular routing...
passed.
Checking reach connections for proximity...
5 segments with non-adjacent reaches found.
At segments:
41 20 21 22 23
5 segments with non-adjacent reaches found.
At segments:
41 20 21 22 23
Checking for model cells with multiple non-zero SFR conductances...
24 model cells with multiple non-zero SFR conductances found.
This may lead to circular routing between collocated reaches.
Nodes with overlapping conductances:
k i j iseg ireach rchlen strthick strhc1
2 110 119 1 8 268.5836486816406 1.0 1.0
1 110 119 2 1 72.61438751220703 1.0 1.0
0 87 122 3 54 35.35249328613281 1.0 1.0
0 87 122 4 33 231.09567260742188 1.0 1.0
0 76 110 5 47 204.88766479492188 1.0 1.0
0 87 122 6 1 30.414600372314453 1.0 1.0
2 84 119 6 7 271.24835205078125 1.0 1.0
2 84 119 6 9 26.37481117248535 1.0 1.0
0 76 110 6 27 75.33216094970703 1.0 1.0
2 25 24 7 42 95.27275085449219 1.0 1.0
2 25 24 8 47 230.57635498046875 1.0 1.0
1 58 132 9 25 48.14010238647461 1.0 1.0
1 58 132 9 27 92.92947387695312 1.0 1.0
0 48 109 9 64 251.07447814941406 1.0 1.0
0 76 110 10 1 69.20076751708984 1.0 1.0
0 48 108 10 40 19.56378746032715 1.0 1.0
0 48 109 10 41 122.84815216064453 1.0 1.0
0 48 108 10 42 28.33331871032715 1.0 1.0
2 25 24 11 1 21.51937484741211 1.0 1.0
2 18 17 11 15 148.96444702148438 1.0 1.0
2 18 17 12 57 76.79739379882812 1.0 1.0
1 80 49 13 70 159.26600646972656 1.0 1.0
0 48 108 14 1 125.24523162841797 1.0 1.0
0 47 98 14 22 67.60189056396484 1.0 1.0
0 47 98 14 24 103.03018188476562 1.0 1.0
0 54 89 14 39 34.45417785644531 1.0 1.0
0 54 89 14 41 60.66257095336914 1.0 1.0
0 53 78 14 57 61.411861419677734 1.0 1.0
0 53 78 14 59 28.438196182250977 1.0 1.0
0 52 76 14 62 63.360740661621094 1.0 1.0
0 52 76 14 64 294.8589782714844 1.0 1.0
1 54 73 14 69 341.6566162109375 1.0 1.0
1 54 73 14 71 27.671581268310547 1.0 1.0
1 80 49 14 125 39.762516021728516 1.0 1.0
2 18 17 16 1 185.54652404785156 1.0 1.0
2 15 3 16 21 32.64251708984375 1.0 1.0
1 36 11 17 38 57.836700439453125 1.0 1.0
2 110 159 19 2 201.63583374023438 1.0 1.0
0 38 85 21 8 72.7103042602539 1.0 1.0
0 38 85 21 9 54.39541244506836 1.0 1.0
2 108 57 24 8 237.04415893554688 1.0 1.0
2 108 57 24 9 46.142940521240234 1.0 1.0
0 94 35 24 45 96.94855499267578 1.0 1.0
0 94 35 24 47 91.54985046386719 1.0 1.0
2 91 35 24 50 60.89990997314453 1.0 1.0
2 91 34 24 51 61.16813278198242 1.0 1.0
1 80 49 25 1 146.93106079101562 1.0 1.0
2 91 35 25 26 308.2864685058594 1.0 1.0
2 91 34 25 27 75.82389068603516 1.0 1.0
2 15 3 31 1 273.603271484375 1.0 1.0
2 36 11 36 1 263.1260681152344 1.0 1.0
2 110 159 38 1 24.2677001953125 1.0 1.0
2 91 34 42 1 130.52822875976562 1.0 1.0
1 92 30 42 6 66.65867614746094 1.0 1.0
1 92 30 42 8 252.91909790039062 1.0 1.0
Checking for streambed tops of less than -10...
passed.
Checking for streambed tops of greater than 15000...
passed.
Checking segment_data for downstream rises in streambed elevation...
Segment elevup and elevdn not specified for nstrm=-1439 and isfropt=1
passed.
Checking reach_data for downstream rises in streambed elevation...
passed.
Checking reach_data for inconsistencies between streambed elevations and the model grid...
186 reaches encountered with streambed above model top.
Model top violations:
i j iseg ireach strtop modeltop strhc1 reachID diff
44 51 8 2 1096.0574951171875 1084.9100341796875 1.0 214 -11.1474609375
31 53 23 10 1008.349853515625 998.0900268554688 1.0 931 -10.25982666015625
34 50 12 5 1016.0361328125 1006.780029296875 1.0 385 -9.256103515625
42 49 8 6 1066.8226318359375 1057.7900390625 1.0 218 -9.0325927734375
3 76 27 3 1009.2495727539062 1002.1099853515625 1.0 1082 -7.13958740234375
94 3 37 21 1073.4644775390625 1066.4599609375 1.0 1360 -7.0045166015625
61 22 18 2 1029.281982421875 1022.3200073242188 1.0 769 -6.96197509765625
84 111 5 34 1261.253662109375 1254.3699951171875 1.0 130 -6.8836669921875
47 29 15 21 965.2784423828125 958.4199829101562 1.0 653 -6.85845947265625
61 18 33 3 996.3890991210938 989.6300048828125 1.0 1246 -6.75909423828125
71 5 35 9 990.4193725585938 983.8200073242188 1.0 1310 -6.599365234375
43 50 8 4 1078.9158935546875 1072.3599853515625 1.0 216 -6.555908203125
84 9 34 2 1085.3753662109375 1079.010009765625 1.0 1287 -6.3653564453125
35 51 12 3 1028.656494140625 1022.8200073242188 1.0 383 -5.83648681640625
108 54 24 12 1228.1346435546875 1222.3299560546875 1.0 1010 -5.8046875
33 4 36 10 834.1530151367188 828.52001953125 1.0 1334 -5.63299560546875
0 100 40 1 1027.534423828125 1022.1199951171875 1.0 1384 -5.4144287109375
5 25 28 45 817.1793823242188 811.8099975585938 1.0 1135 -5.369384765625
22 12 15 62 824.2842407226562 819.0800170898438 1.0 694 -5.2042236328125
52 37 15 9 1054.0181884765625 1048.8399658203125 1.0 641 -5.17822265625
65 1 35 19 944.7957153320312 939.6599731445312 1.0 1320 -5.1357421875
49 15 18 19 925.3494262695312 920.3099975585938 1.0 786 -5.0394287109375
51 24 17 11 963.3839721679688 958.3800048828125 1.0 740 -5.00396728515625
26 47 23 21 942.5027465820312 937.5900268554688 1.0 942 -4.9127197265625
1 23 28 51 778.961669921875 774.219970703125 1.0 1141 -4.74169921875
73 5 35 7 1003.2875366210938 998.5999755859375 1.0 1308 -4.68756103515625
45 28 15 24 947.1143188476562 942.72998046875 1.0 656 -4.38433837890625
80 116 6 18 1229.7757568359375 1225.4599609375 1.0 161 -4.3157958984375
27 18 15 51 863.89599609375 859.6300048828125 1.0 683 -4.2659912109375
82 112 5 39 1242.8052978515625 1238.5400390625 1.0 135 -4.2652587890625
35 31 7 25 922.0911254882812 918.2000122070312 1.0 195 -3.89111328125
30 52 23 12 991.4169921875 987.5700073242188 1.0 933 -3.84698486328125
71 4 35 10 981.303466796875 977.6799926757812 1.0 1311 -3.62347412109375
58 19 18 6 986.8626708984375 983.260009765625 1.0 773 -3.6026611328125
87 110 5 30 1283.4881591796875 1279.989990234375 1.0 126 -3.4981689453125
59 17 33 5 976.4671630859375 973.0399780273438 1.0 1248 -3.42718505859375
85 111 5 33 1264.7679443359375 1261.3499755859375 1.0 129 -3.41796875
41 12 18 31 892.8302001953125 889.469970703125 1.0 798 -3.3602294921875
54 12 33 15 932.0604248046875 928.719970703125 1.0 1258 -3.3404541015625
16 32 23 46 867.0645751953125 863.75 1.0 967 -3.3145751953125
28 40 12 21 934.5240478515625 931.27001953125 1.0 401 -3.2540283203125
1 138 26 1 1154.71630859375 1151.489990234375 1.0 1077 -3.226318359375
10 25 23 59 824.5831909179688 821.3699951171875 1.0 980 -3.21319580078125
36 32 7 23 927.737060546875 924.5399780273438 1.0 193 -3.19708251953125
51 15 18 17 930.7290649414062 927.5700073242188 1.0 784 -3.1590576171875
67 2 35 16 953.3693237304688 950.25 1.0 1317 -3.11932373046875
82 140 3 27 1305.33447265625 1302.239990234375 1.0 36 -3.094482421875
58 4 32 22 912.39599609375 909.3099975585938 1.0 1234 -3.08599853515625
31 28 7 32 902.4729614257812 899.4500122070312 1.0 202 -3.02294921875
18 0 30 40 762.85107421875 759.8400268554688 1.0 1204 -3.01104736328125
88 110 5 29 1288.3612060546875 1285.3800048828125 1.0 125 -2.981201171875
34 30 7 27 917.8261108398438 914.8599853515625 1.0 197 -2.96612548828125
33 13 30 13 879.4591674804688 876.5599975585938 1.0 1177 -2.899169921875
1 137 26 2 1149.9388427734375 1147.0799560546875 1.0 1078 -2.85888671875
109 58 24 6 1244.657470703125 1241.8199462890625 1.0 1004 -2.8375244140625
6 22 23 66 783.7332153320312 780.9299926757812 1.0 987 -2.80322265625
28 7 30 24 838.6741943359375 835.8900146484375 1.0 1188 -2.7841796875
83 51 13 65 1143.177490234375 1140.4300537109375 1.0 502 -2.7474365234375
20 9 15 67 797.5311279296875 794.7999877929688 1.0 699 -2.73114013671875
83 145 3 21 1313.221923828125 1310.52001953125 1.0 30 -2.701904296875
34 43 8 20 971.7984619140625 969.0999755859375 1.0 232 -2.698486328125
43 39 7 10 1002.135009765625 999.4600219726562 1.0 180 -2.67498779296875
18 16 16 2 816.8546142578125 814.1799926757812 1.0 710 -2.67462158203125
80 2 34 13 1005.6331176757812 1002.989990234375 1.0 1298 -2.64312744140625
57 27 17 2 1030.468994140625 1027.8399658203125 1.0 731 -2.6290283203125
98 6 37 14 1130.2991943359375 1127.739990234375 1.0 1353 -2.5592041015625
96 6 37 16 1109.81494140625 1107.27001953125 1.0 1355 -2.544921875
41 27 15 29 922.3822631835938 919.8499755859375 1.0 661 -2.53228759765625
93 35 24 48 1133.5345458984375 1131.030029296875 1.0 1046 -2.5045166015625
11 26 23 57 835.0635986328125 832.5999755859375 1.0 978 -2.463623046875
57 18 18 8 976.854248046875 974.4600219726562 1.0 775 -2.39422607421875
43 27 15 27 935.5574951171875 933.1699829101562 1.0 659 -2.38751220703125
29 19 15 48 875.1234130859375 872.77001953125 1.0 680 -2.3533935546875
97 6 37 15 1122.60205078125 1120.280029296875 1.0 1354 -2.322021484375
76 110 6 27 1202.379638671875 1200.0799560546875 1.0 170 -2.2996826171875
13 29 23 52 853.8441162109375 851.5800170898438 1.0 973 -2.26409912109375
28 50 23 16 969.1011352539062 966.8800048828125 1.0 937 -2.22113037109375
41 0 33 40 858.0049438476562 855.7899780273438 1.0 1283 -2.2149658203125
25 16 15 55 848.8750610351562 846.7100219726562 1.0 687 -2.1650390625
3 24 28 48 797.5549926757812 795.4099731445312 1.0 1138 -2.14501953125
65 10 32 9 973.0398559570312 970.9299926757812 1.0 1221 -2.10986328125
98 100 5 9 1381.1871337890625 1379.1099853515625 1.0 105 -2.0771484375
21 25 12 43 869.5907592773438 867.52001953125 1.0 423 -2.07073974609375
78 111 5 44 1220.5587158203125 1218.530029296875 1.0 140 -2.0286865234375
55 25 17 6 996.66845703125 994.6400146484375 1.0 735 -2.0284423828125
51 36 15 11 1035.9466552734375 1033.9599609375 1.0 643 -1.9866943359375
30 34 8 32 918.734375 916.7899780273438 1.0 244 -1.94439697265625
32 21 15 44 887.8782348632812 885.9400024414062 1.0 676 -1.938232421875
4 77 27 1 1019.9188232421875 1018.0 1.0 1080 -1.9188232421875
51 9 33 21 916.0432739257812 914.1300048828125 1.0 1264 -1.91326904296875
27 6 30 26 830.7896118164062 828.9600219726562 1.0 1190 -1.82958984375
6 26 28 43 833.5056762695312 831.719970703125 1.0 1133 -1.78570556640625
22 2 30 34 786.0257568359375 784.25 1.0 1198 -1.7757568359375
7 26 28 42 844.1419677734375 842.3800048828125 1.0 1132 -1.761962890625
48 6 33 27 898.0303955078125 896.2899780273438 1.0 1270 -1.74041748046875
18 21 12 51 847.4663696289062 845.739990234375 1.0 431 -1.72637939453125
89 64 13 41 1181.4326171875 1179.72998046875 1.0 478 -1.70263671875
74 6 35 5 1011.9109497070312 1010.219970703125 1.0 1306 -1.69097900390625
8 69 22 4 1005.525390625 1003.8599853515625 1.0 904 -1.6654052734375
13 45 28 18 909.0931396484375 907.5 1.0 1108 -1.5931396484375
43 13 18 28 900.6517333984375 899.0599975585938 1.0 795 -1.59173583984375
46 14 18 24 910.212890625 908.7100219726562 1.0 791 -1.50286865234375
15 6 16 16 769.787109375 768.2899780273438 1.0 724 -1.49713134765625
3 39 29 12 893.4083862304688 891.9400024414062 1.0 1155 -1.4683837890625
84 145 3 20 1316.32958984375 1314.9100341796875 1.0 29 -1.4195556640625
12 28 23 54 846.7885131835938 845.3699951171875 1.0 975 -1.41851806640625
20 1 30 37 775.9326171875 774.530029296875 1.0 1201 -1.402587890625
47 5 33 29 893.9849243164062 892.5900268554688 1.0 1272 -1.3948974609375
57 3 32 24 906.1943359375 904.8900146484375 1.0 1236 -1.3043212890625
37 20 30 2 917.4218139648438 916.1199951171875 1.0 1166 -1.30181884765625
76 110 5 47 1201.3443603515625 1200.0799560546875 1.0 143 -1.264404296875
76 110 10 1 1201.3443603515625 1200.0799560546875 1.0 324 -1.264404296875
68 3 35 14 962.5520629882812 961.3599853515625 1.0 1315 -1.19207763671875
51 119 9 45 1191.214111328125 1190.050048828125 1.0 304 -1.1640625
25 46 23 23 935.9991455078125 934.8499755859375 1.0 944 -1.149169921875
39 26 15 32 915.0709838867188 913.97998046875 1.0 664 -1.09100341796875
70 3 35 12 969.0398559570312 967.9500122070312 1.0 1313 -1.08984375
6 158 39 3 1123.38037109375 1122.300048828125 1.0 1372 -1.080322265625
37 25 15 35 908.1263427734375 907.0700073242188 1.0 667 -1.05633544921875
26 25 7 40 879.2858276367188 878.239990234375 1.0 210 -1.04583740234375
97 41 24 35 1150.635498046875 1149.6500244140625 1.0 1033 -0.9854736328125
60 107 10 26 1175.4578857421875 1174.52001953125 1.0 349 -0.9378662109375
46 21 17 19 923.8571166992188 922.9199829101562 1.0 748 -0.9371337890625
25 158 20 3 1172.1849365234375 1171.27001953125 1.0 807 -0.9149169921875
39 48 8 10 1029.3406982421875 1028.449951171875 1.0 222 -0.8907470703125
14 99 21 61 1097.334716796875 1096.469970703125 1.0 879 -0.86474609375
40 86 21 5 1143.69384765625 1142.8499755859375 1.0 823 -0.8438720703125
85 52 13 62 1149.013427734375 1148.1700439453125 1.0 499 -0.8433837890625
52 109 10 35 1164.787353515625 1163.969970703125 1.0 358 -0.8173828125
38 12 17 36 880.8272094726562 880.030029296875 1.0 765 -0.79718017578125
74 8 35 3 1025.747314453125 1024.9599609375 1.0 1304 -0.787353515625
105 51 24 18 1209.4859619140625 1208.699951171875 1.0 1016 -0.7860107421875
51 108 10 37 1162.79296875 1162.010009765625 1.0 360 -0.782958984375
25 33 12 31 903.5383911132812 902.760009765625 1.0 411 -0.77838134765625
28 25 7 38 886.2448120117188 885.5 1.0 208 -0.74481201171875
0 71 27 11 967.5951538085938 966.8599853515625 1.0 1090 -0.73516845703125
66 10 32 8 977.9095458984375 977.219970703125 1.0 1220 -0.6895751953125
5 157 39 5 1121.365966796875 1120.6800537109375 1.0 1374 -0.6859130859375
45 13 18 26 906.5974731445312 905.9299926757812 1.0 793 -0.66748046875
26 158 20 2 1174.694580078125 1174.030029296875 1.0 806 -0.66455078125
62 19 33 1 1005.5029296875 1004.8400268554688 1.0 1244 -0.66290283203125
56 18 18 9 967.2000732421875 966.5399780273438 1.0 776 -0.66009521484375
33 80 21 19 1128.41845703125 1127.780029296875 1.0 837 -0.638427734375
101 41 24 31 1158.5797119140625 1157.969970703125 1.0 1029 -0.6097412109375
74 109 10 4 1198.0435791015625 1197.43994140625 1.0 327 -0.6036376953125
30 10 30 19 857.75048828125 857.1599731445312 1.0 1183 -0.59051513671875
96 36 24 42 1141.318359375 1140.72998046875 1.0 1040 -0.58837890625
33 22 15 42 892.9962768554688 892.4299926757812 1.0 674 -0.5662841796875
6 104 21 74 1082.59326171875 1082.06005859375 1.0 892 -0.533203125
104 44 24 26 1173.2772216796875 1172.8199462890625 1.0 1024 -0.457275390625
42 27 15 28 927.3306884765625 926.8800048828125 1.0 660 -0.45068359375
26 17 15 53 855.5484008789062 855.1099853515625 1.0 685 -0.43841552734375
93 122 4 27 1282.7025146484375 1282.2900390625 1.0 90 -0.4124755859375
20 89 21 45 1108.5118408203125 1108.0999755859375 1.0 863 -0.411865234375
32 29 7 30 907.5672607421875 907.1599731445312 1.0 200 -0.40728759765625
6 44 29 4 923.0087890625 922.6599731445312 1.0 1147 -0.34881591796875
98 41 24 34 1152.4619140625 1152.1199951171875 1.0 1032 -0.3419189453125
60 6 32 18 933.5938720703125 933.260009765625 1.0 1230 -0.3338623046875
37 21 30 1 919.2725830078125 918.9400024414062 1.0 1165 -0.33258056640625
89 122 4 31 1278.8204345703125 1278.5 1.0 94 -0.3204345703125
57 108 10 30 1171.8583984375 1171.5400390625 1.0 353 -0.318359375
34 81 21 17 1129.405517578125 1129.0899658203125 1.0 835 -0.3155517578125
82 50 13 67 1139.371337890625 1139.06005859375 1.0 504 -0.311279296875
53 108 10 34 1166.6656494140625 1166.4000244140625 1.0 357 -0.265625
88 55 13 55 1161.44287109375 1161.199951171875 1.0 492 -0.242919921875
47 105 14 7 1156.3297119140625 1156.0999755859375 1.0 514 -0.229736328125
58 108 10 29 1172.513916015625 1172.2900390625 1.0 352 -0.223876953125
99 123 4 18 1290.2703857421875 1290.0999755859375 1.0 81 -0.17041015625
101 95 5 1 1400.4940185546875 1400.3399658203125 1.0 97 -0.154052734375
36 19 30 4 910.9712524414062 910.8200073242188 1.0 1168 -0.1512451171875
86 52 13 61 1151.74169921875 1151.5999755859375 1.0 498 -0.1417236328125
23 2 30 33 790.0592651367188 789.9199829101562 1.0 1197 -0.1392822265625
36 46 8 15 997.1134033203125 996.97998046875 1.0 227 -0.1334228515625
15 31 23 48 859.490234375 859.3599853515625 1.0 969 -0.1302490234375
16 3 15 76 757.1549682617188 757.0399780273438 1.0 708 -0.114990234375
99 14 37 5 1152.94384765625 1152.8499755859375 1.0 1344 -0.0938720703125
89 56 13 53 1164.082763671875 1163.989990234375 1.0 490 -0.0927734375
91 122 4 29 1280.729736328125 1280.6400146484375 1.0 92 -0.0897216796875
35 58 23 1 1069.680419921875 1069.5999755859375 1.0 922 -0.0804443359375
52 81 14 51 1142.7772216796875 1142.699951171875 1.0 558 -0.0772705078125
46 120 9 51 1181.7955322265625 1181.719970703125 1.0 310 -0.0755615234375
40 48 8 9 1038.634765625 1038.56005859375 1.0 221 -0.07470703125
99 41 24 33 1153.8238525390625 1153.75 1.0 1031 -0.0738525390625
75 110 10 2 1199.30908203125 1199.25 1.0 325 -0.05908203125
47 120 9 50 1182.4256591796875 1182.3699951171875 1.0 309 -0.0556640625
98 123 4 19 1289.3104248046875 1289.2900390625 1.0 82 -0.0203857421875
Checking segment_data for inconsistencies between segment end elevations and the model grid...
Segment elevup and elevdn not specified for nstrm=-1439 and isfropt=1
passed.
Checking for streambed slopes of less than 0.0001...
passed.
Checking for streambed slopes of greater than 1.0...
passed.
wrote tf_SFR.chk
converting segment data to period data...
converting reach and segment data to package data...
wrote tf_mf6.sfr
Writing tables
The reach_data and segment_data tables can be written out to csv files for further processing:
[21]:
sfrdata.write_tables()
wrote tables/tf_sfr_reach_data.csv
wrote tables/tf_sfr_segment_data.csv
and then read back in to create an SFRData instance
[22]:
sfrdata2 = sfrmaker.SFRData.from_tables('tables/tf_sfr_reach_data.csv', 'tables/tf_sfr_segment_data.csv',
grid=grid)
Model grid information
structured grid
nnodes: 17,760
nlay: 1
nrow: 111
ncol: 160
model length units: undefined
crs: EPSG:26715
bounds: 682688.00, 5139052.00, 694880.00, 5147510.20
active area defined by: isfr array
Writing shapefiles
Shapefiles can also be written to visualize the SFR package. These include:
hydrography linestrings associated with each reach
model cell polygons associated with each reach
outlet locations where water is leaving the model
routing connections (as lines drawn between cell centers)
the locations of specified inflows or any other period data
observation locations (if observations were supplied as input)
[23]:
sfrdata.write_shapefiles()
writing shps/tf_sfr_cells.shp... Done
writing shps/tf_sfr_outlets.shp... Done
writing shps/tf_sfr_lines.shp... Done
writing shps/tf_sfr_routing.shp... Done
No period data to export!
No observations to export!
No non-zero values of flow to export!
[ ]: