Note
Go to the end to download the full example code.
1D Rectilinear Mesh
from copy import deepcopy
from geobipy import DataArray, StatArray
from geobipy import RectilinearMesh1D, RectilinearMesh2D, RectilinearMesh2D_stitched
import matplotlib.gridspec as gridspec
import matplotlib.pyplot as plt
import numpy as np
import h5py
The basics
Instantiate a new 1D rectilinear mesh by specifying cell centres, edges, or widths.
x = StatArray(np.cumsum(np.arange(0.0, 10.0)), 'Depth', 'm')
Cell edges
rm = RectilinearMesh1D(edges=x, centres=None, widths=None)
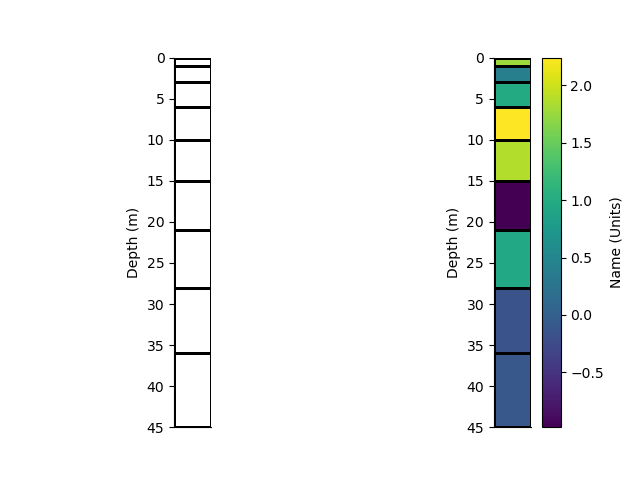
We can plot the grid of the mesh Or Pcolor the mesh showing. An array of cell values is used as the colour.
arr = StatArray(np.random.randn(*rm.shape), "Name", "Units")
p=0; plt.figure(p)
plt.subplot(121)
_ = rm.plot_grid(transpose=True, flip=True)
plt.subplot(122)
_ = rm.pcolor(arr, grid=True, transpose=True, flip=True)
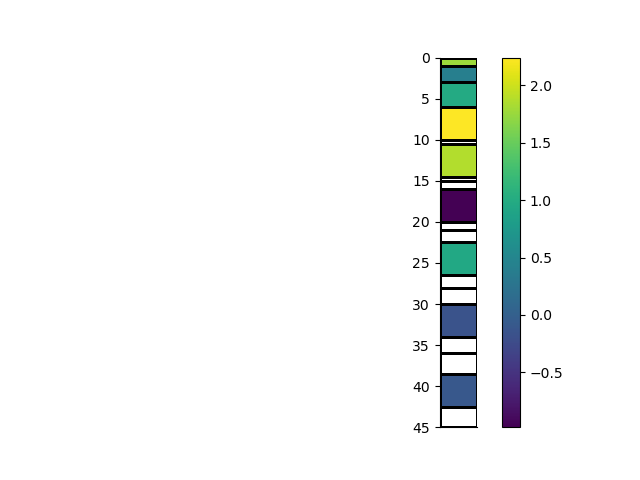
# Mask the mesh cells by a distance
rm_masked, indices, arr2 = rm.mask_cells(2.0, values=arr)
p+=1; plt.figure(p)
_ = rm_masked.pcolor(StatArray(arr2), grid=True, transpose=True, flip=True)
# Writing and reading to/from HDF5
# ++++++++++++++++++++++++++++++++
with h5py.File('rm1d.h5', 'w') as f:
rm.toHdf(f, 'rm1d')
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'])
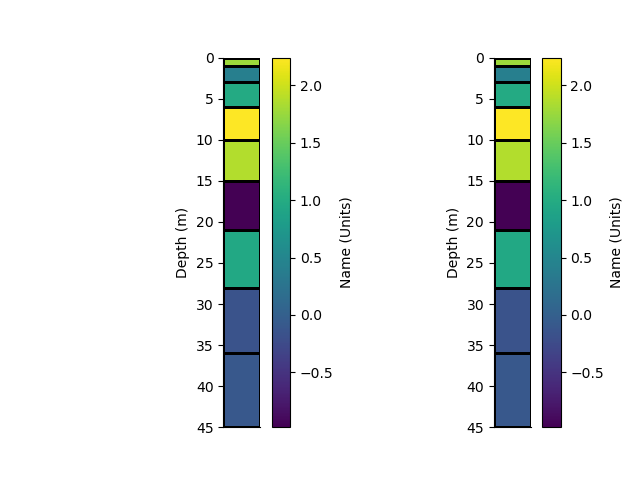
p+=1; plt.figure(p)
plt.subplot(121)
_ = rm.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
plt.subplot(122)
_ = rm1.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
with h5py.File('rm1d.h5', 'w') as f:
rm.createHdf(f, 'rm1d', add_axis=10)
for i in range(10):
rm.writeHdf(f, 'rm1d', index=i)
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'], index=0)
with h5py.File('rm1d.h5', 'r') as f:
rm2 = RectilinearMesh2D.fromHdf(f['rm1d'])
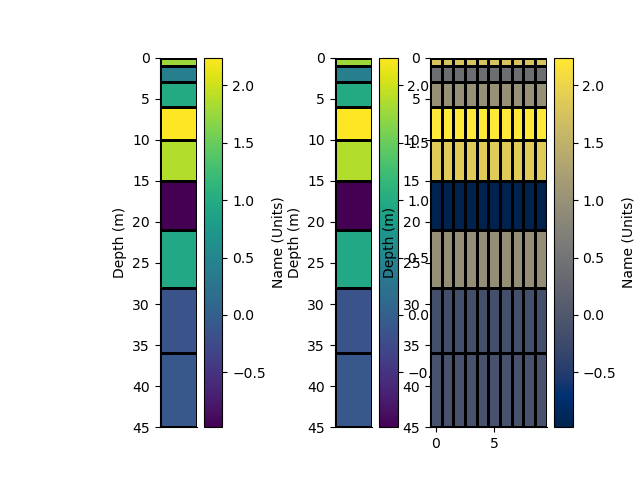
p+=1; plt.figure(p)
plt.subplot(131)
_ = rm.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
plt.subplot(132)
_ = rm1.pcolor(arr, grid=True, transpose=True, flip=True)
plt.subplot(133)
_ = rm2.pcolor(np.repeat(arr[None, :], 10, 0), grid=True, flipY=True)
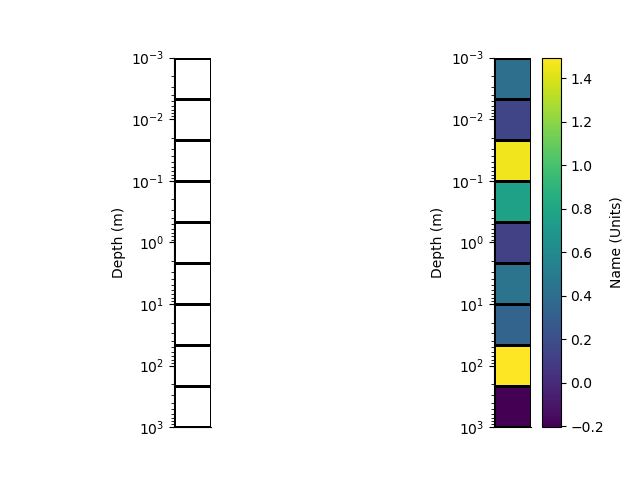
Log-space rectilinear mesh
Instantiate a new 1D rectilinear mesh by specifying cell centres or edges. Here we use edges
x = StatArray(np.logspace(-3, 3, 10), 'Depth', 'm')
rm = RectilinearMesh1D(edges=x, log=10)
# We can plot the grid of the mesh
# Or Pcolor the mesh showing. An array of cell values is used as the colour.
p+=1; plt.figure(p)
plt.subplot(121)
_ = rm.plot_grid(transpose=True, flip=True)
plt.subplot(122)
arr = StatArray(np.random.randn(rm.nCells), "Name", "Units")
_ = rm.pcolor(arr, grid=True, transpose=True, flip=True)
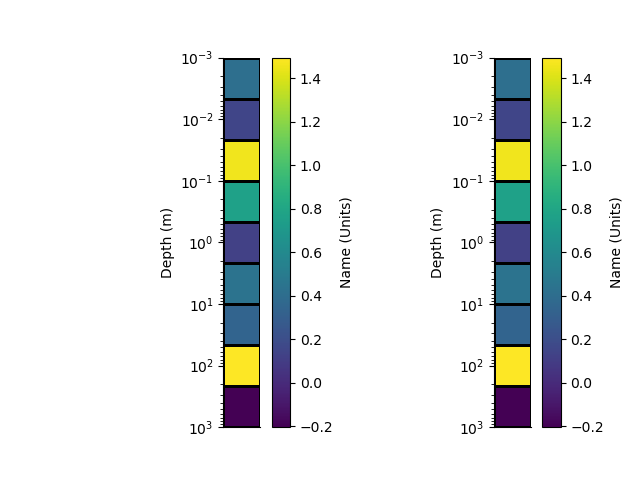
# Writing and reading to/from HDF5
# ++++++++++++++++++++++++++++++++
with h5py.File('rm1d.h5', 'w') as f:
rm.toHdf(f, 'rm1d')
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'])
p+=1; plt.figure(p)
plt.subplot(121)
_ = rm.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
plt.subplot(122)
_ = rm1.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
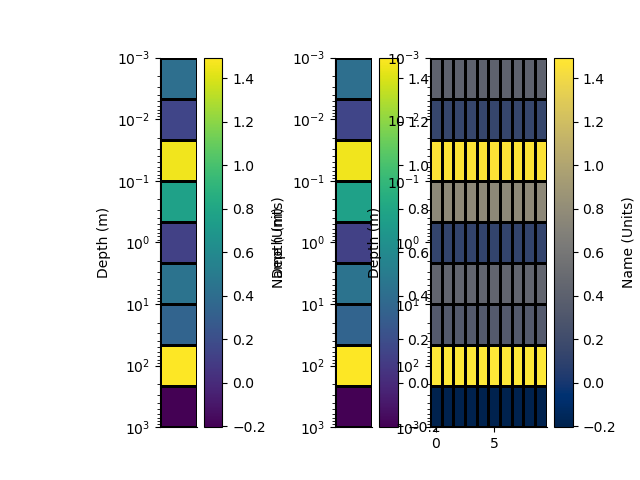
with h5py.File('rm1d.h5', 'w') as f:
rm.createHdf(f, 'rm1d', add_axis=10)
for i in range(10):
rm.writeHdf(f, 'rm1d', index=i)
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'], index=0)
with h5py.File('rm1d.h5', 'r') as f:
rm2 = RectilinearMesh2D.fromHdf(f['rm1d'])
p+=1; plt.figure(p)
plt.subplot(131)
_ = rm.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
plt.subplot(132)
_ = rm1.pcolor(arr, grid=True, transpose=True, flip=True)
plt.subplot(133)
_ = rm2.pcolor(np.repeat(arr[None, :], 10, 0), grid=True, flipY=True)
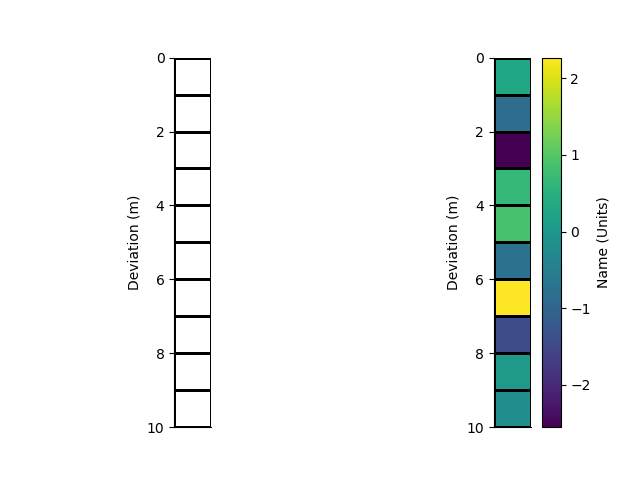
relative_to
Instantiate a new 1D rectilinear mesh by specifying cell centres or edges. Here we use edges
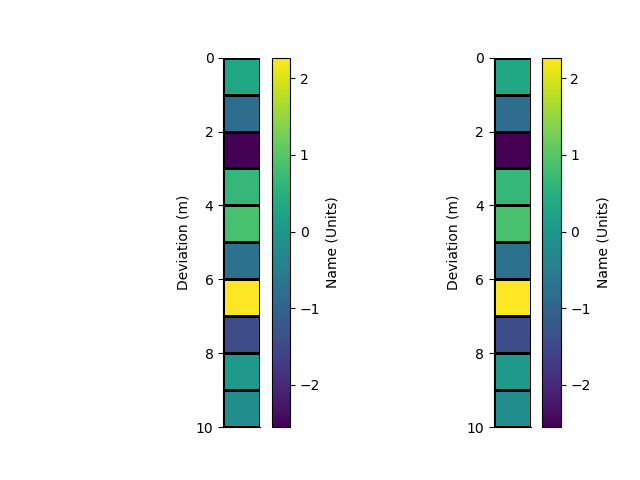
x = StatArray(np.arange(11.0), 'Deviation', 'm')
rm = RectilinearMesh1D(edges=x, relative_to=5.0)
We can plot the grid of the mesh Or Pcolor the mesh showing. An array of cell values is used as the colour.
p+=1; plt.figure(p)
plt.subplot(121)
_ = rm.plot_grid(transpose=True, flip=True)
plt.subplot(122)
arr = StatArray(np.random.randn(rm.nCells), "Name", "Units")
_ = rm.pcolor(arr, grid=True, transpose=True, flip=True)
# Writing and reading to/from HDF5
# ++++++++++++++++++++++++++++++++
with h5py.File('rm1d.h5', 'w') as f:
rm.createHdf(f, 'rm1d')
rm.writeHdf(f, 'rm1d')
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'])
p+=1; plt.figure(p)
plt.subplot(121)
_ = rm.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
plt.subplot(122)
_ = rm1.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
with h5py.File('rm1d.h5', 'w') as f:
rm.createHdf(f, 'rm1d', add_axis=3)
for i in range(3):
rm.relative_to += 0.5
rm.writeHdf(f, 'rm1d', index=i)
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'], index=0)
with h5py.File('rm1d.h5', 'r') as f:
rm2 = RectilinearMesh2D.fromHdf(f['rm1d'])
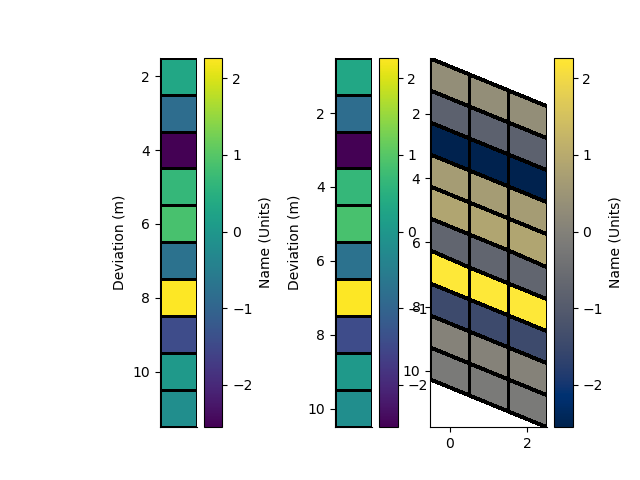
p+=1; plt.figure(p)
plt.subplot(131)
_ = rm.pcolor(StatArray(arr), grid=True, transpose=True, flip=True)
plt.subplot(132)
_ = rm1.pcolor(arr, grid=True, transpose=True, flip=True)
plt.subplot(133)
_ = rm2.pcolor(np.repeat(arr[None, :], 3, 0), grid=True, flipY=True)
# Making a mesh perturbable
# +++++++++++++++++++++++++
n_cells = 2
widths = DataArray(np.full(n_cells, fill_value=10.0), 'test')
rm = RectilinearMesh1D(widths=widths, relative_to=0.0)
Randomness and Model Perturbations
We can set the priors on the 1D model by assigning minimum and maximum layer depths and a maximum number of layers. These are used to create priors on the number of cells in the model, a new depth interface, new parameter values and the vertical gradient of those parameters. The halfSpaceValue is used as a reference value for the parameter prior.
from numpy.random import Generator
from numpy.random import PCG64DXSM
generator = PCG64DXSM(seed=0)
prng = Generator(generator)
# Set the priors
rm.set_priors(min_edge = 1.0,
max_edge = 150.0,
max_cells = 30,
prng = prng)
We can evaluate the prior of the model using depths only
print('Log probability of the Mesh given its priors: ', rm.probability)
Log probability of the Mesh given its priors: -3.367295829986474
To propose new meshes, we specify the probabilities of creating, removing, perturbing, and not changing an edge interface Here we force the creation of a layer.
rm.set_proposals(probabilities = [0.25, 0.25, 0.25, 0.25], prng=prng)
rm.set_posteriors()
rm0 = deepcopy(rm)
We can then perturb the layers of the model
for i in range(1000):
rm = rm.perturb()
rm.update_posteriors()
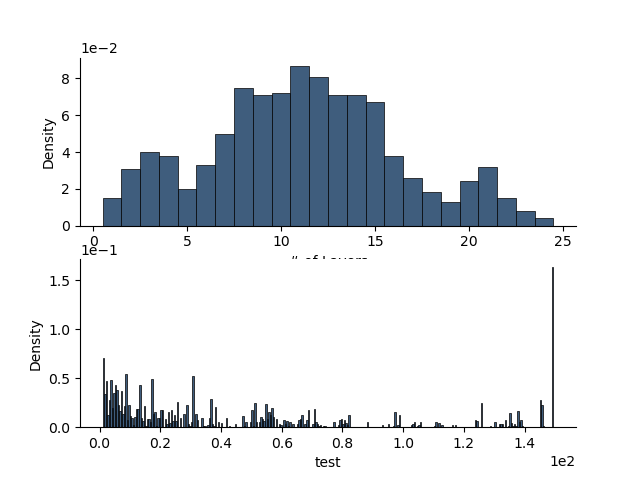
p+=1; fig = plt.figure(p)
ax = rm._init_posterior_plots(fig)
rm.plot_posteriors(axes=ax)
with h5py.File('rm1d.h5', 'w') as f:
rm.createHdf(f, 'rm1d', withPosterior = True)
rm.writeHdf(f, 'rm1d', withPosterior = True)
with h5py.File('rm1d.h5', 'r') as f:
rm1 = RectilinearMesh1D.fromHdf(f['rm1d'])
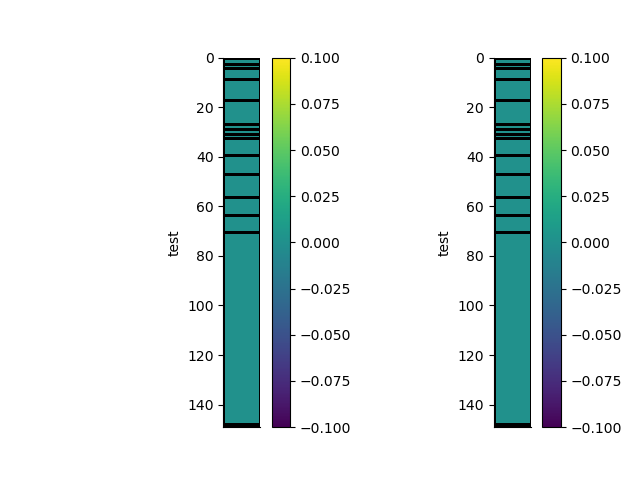
p+=1; plt.figure(p)
plt.subplot(121)
_ = rm.pcolor(StatArray(rm.shape), grid=True, transpose=True, flip=True)
plt.subplot(122)
_ = rm1.pcolor(StatArray(rm1.shape), grid=True, transpose=True, flip=True)
p+=1; fig = plt.figure(p)
ax = rm1._init_posterior_plots(fig)
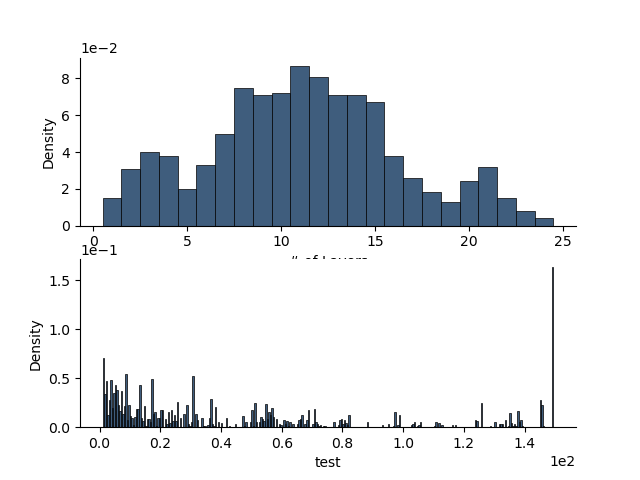
rm1.plot_posteriors(axes=ax)
[<Axes: xlabel='# of Layers', ylabel='Density'>, <Axes: xlabel='test', ylabel='Density'>]
Expanded
with h5py.File('rm1d.h5', 'w') as f:
tmp = rm.pad(rm.max_cells)
tmp.createHdf(f, 'rm1d', withPosterior=True, add_axis=DataArray(np.arange(3.0), name='Easting', units="m"))
print(list(f['rm1d'].keys()))
rm.relative_to = 5.0
print(rm.summary)
rm.writeHdf(f, 'rm1d', withPosterior = True, index=0)
rm = deepcopy(rm0)
for i in range(1000):
rm = rm.perturb(); rm.update_posteriors()
rm.relative_to = 10.0
rm.writeHdf(f, 'rm1d', withPosterior = True, index=1)
rm = deepcopy(rm0)
for i in range(1000):
rm = rm.perturb(); rm.update_posteriors()
rm.relative_to = 25.0
rm.writeHdf(f, 'rm1d', withPosterior = True, index=2)
with h5py.File('rm1d.h5', 'r') as f:
rm2 = RectilinearMesh2D.fromHdf(f['rm1d'])
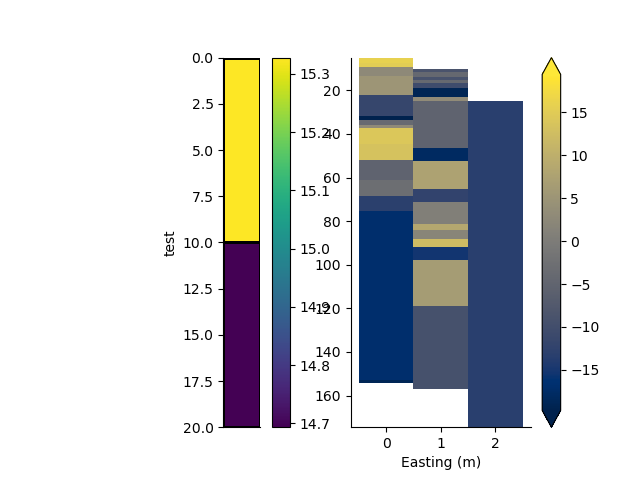
p+=1; plt.figure(p)
plt.subplot(121)
arr = np.random.randn(3, rm.max_cells) * 10
_ = rm0.pcolor(arr[0, :rm0.nCells.item()], grid=True, transpose=True, flip=True)
plt.subplot(122)
_ = rm2.pcolor(arr, grid=True, flipY=True, equalize=True)
from geobipy import RectilinearMesh2D
with h5py.File('rm1d.h5', 'r') as f:
rm2 = RectilinearMesh2D.fromHdf(f['rm1d'], index=0)
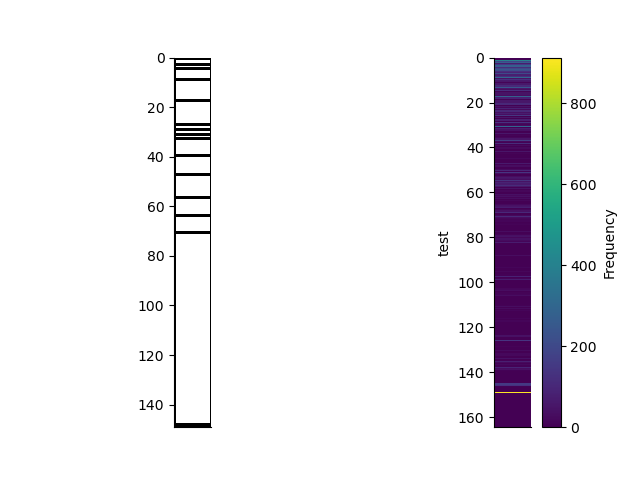
plt.figure()
plt.subplot(121)
rm2.plot_grid(transpose=True, flip=True)
plt.subplot(122)
rm2.edges.posterior.pcolor(transpose=True, flip=True)
plt.show()
['nCells', 'x', 'y']
RectilinearMesh1D
Number of Cells:
| StatArray
| Name: Number of cells
| Address:['0x15dfbb9d0' '0x15d9b10a0' '0x15e28ec10' '0x15e28dc50']
| Shape: (1,)
| Values: [15]
| Min: 15
| Max: 15
| Prior:
| | Uniform Distribution:
| | Min: 1
| | Max: 30
| has_posterior: True
Cell Centres:
| StatArray
| Name: test
| Address:['0x15dc850d0']
| Shape: (15,)
| Values: [ 1.22331766 3.28906551 6.36608642 ... 66.85590133 108.99206549
| 148.449825 ]
| Min: 1.2233176602046558
| Max: 148.4498250024555
| has_posterior: False
Cell Edges:
| StatArray
| Name: test
| Address:['0x155de5e50' '0x15d8381a0' '0x15e28f7b0' '0x153d89c10' '0x15d811b50'
| '0x15d810af0']
| Shape: (16,)
| Values: [ 0. 2.44663532 4.13149571 ... 70.19008745 147.79404353
| 149.10560647]
| Min: 0.0
| Max: 149.1056064716983
| Prior:
| | Order Statistics:
| | None
| Proposal:
| | Uniform Distribution:
| | Min: 1.0
| | Max: 149.99999999999997
| has_posterior: True
log:
| None
relative_to:
| StatArray
| Name:
| Address:['0x15e0e7ed0']
| Shape: (1,)
| Values: [5.]
| Min: 5.0
| Max: 5.0
| has_posterior: False
Total running time of the script: (0 minutes 4.760 seconds)