Determines Pfafstetter codes for a dendritic network with total drainage area, levelpath, and topo_sort attributes.
get_pfaf(x, max_level = 2, status = FALSE)Arguments
Value
data.frame with ID and pfaf columns.
Examples
# \donttest{
library(dplyr)
source(system.file("extdata/nhdplushr_data.R", package = "nhdplusTools"))
hr_flowline <- align_nhdplus_names(hr_data$NHDFlowline)
fl <- select(hr_flowline, COMID, AreaSqKM) %>%
right_join(prepare_nhdplus(hr_flowline, 0, 0,
purge_non_dendritic = FALSE,
warn = FALSE),
by = "COMID") %>%
sf::st_sf() %>%
select(ID = COMID, toID = toCOMID, area = AreaSqKM)
fl$nameID = ""
fl$totda <- calculate_total_drainage_area(sf::st_set_geometry(fl, NULL))
#> Warning: calculate_total_drainage_area() is deprecated. Please switch to hydroloom equivalent.
#> Dendritic routing will be applied. Diversions are assumed to have 0 flow fraction.
fl <- left_join(fl, get_levelpaths(rename(sf::st_set_geometry(fl, NULL),
weight = totda)), by = "ID")
#> Warning: get_levelpaths is deprecated in favor of add_levelpaths in the hydroloom package.
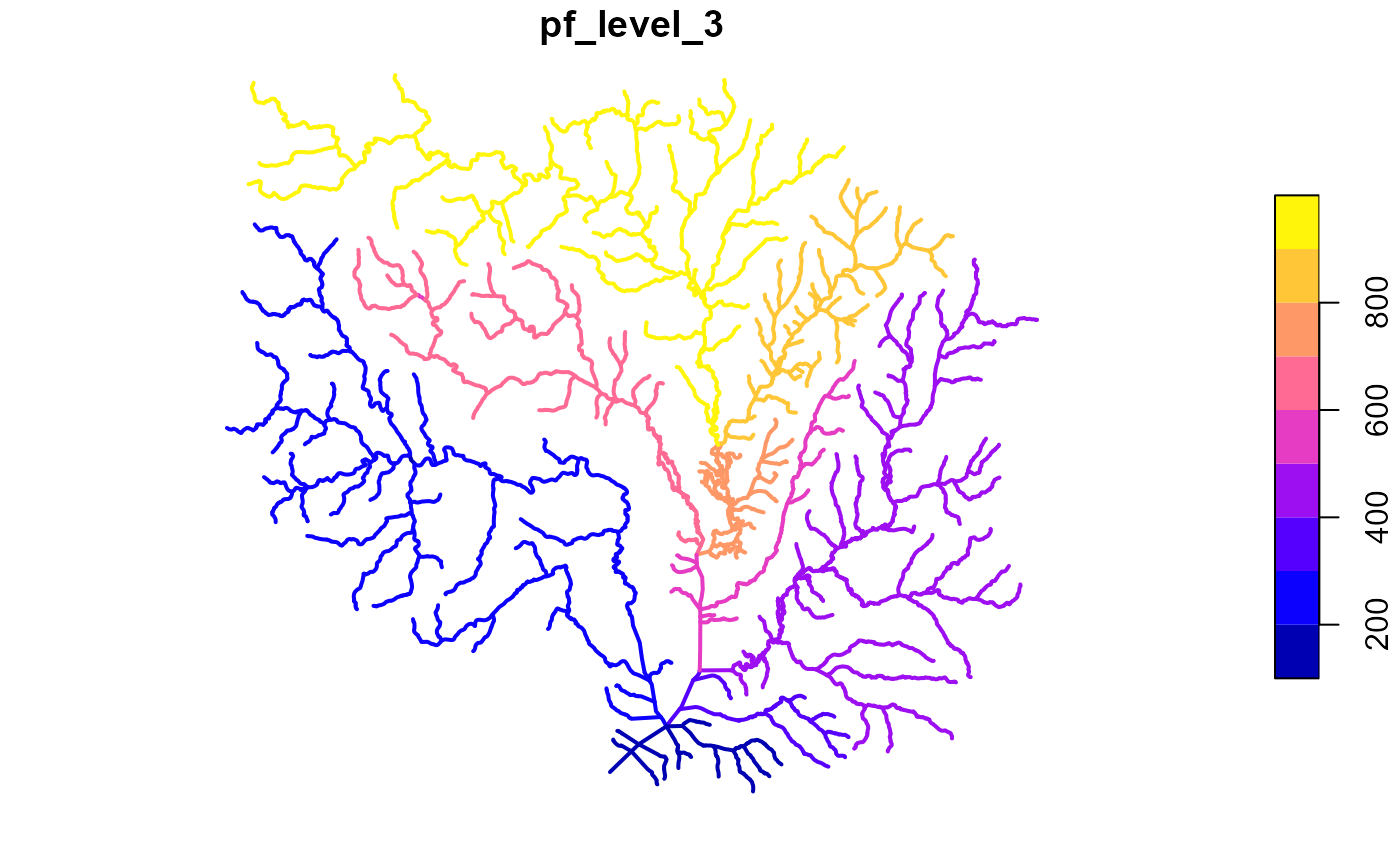
pfaf <- get_pfaf(fl, max_level = 3)
#> Warning: get_pfaf is deprecated, please use hydroloom
fl <- left_join(fl, pfaf, by = "ID")
plot(fl["pf_level_3"], lwd = 2)
 pfaf <- get_pfaf(fl, max_level = 4)
#> Warning: get_pfaf is deprecated, please use hydroloom
hr_catchment <- left_join(hr_data$NHDPlusCatchment, pfaf, by = c("FEATUREID" = "ID"))
colors <- data.frame(pf_level_4 = unique(hr_catchment$pf_level_4),
color = sample(terrain.colors(length(unique(hr_catchment$pf_level_4)))),
stringsAsFactors = FALSE)
hr_catchment <- left_join(hr_catchment, colors, by = "pf_level_4")
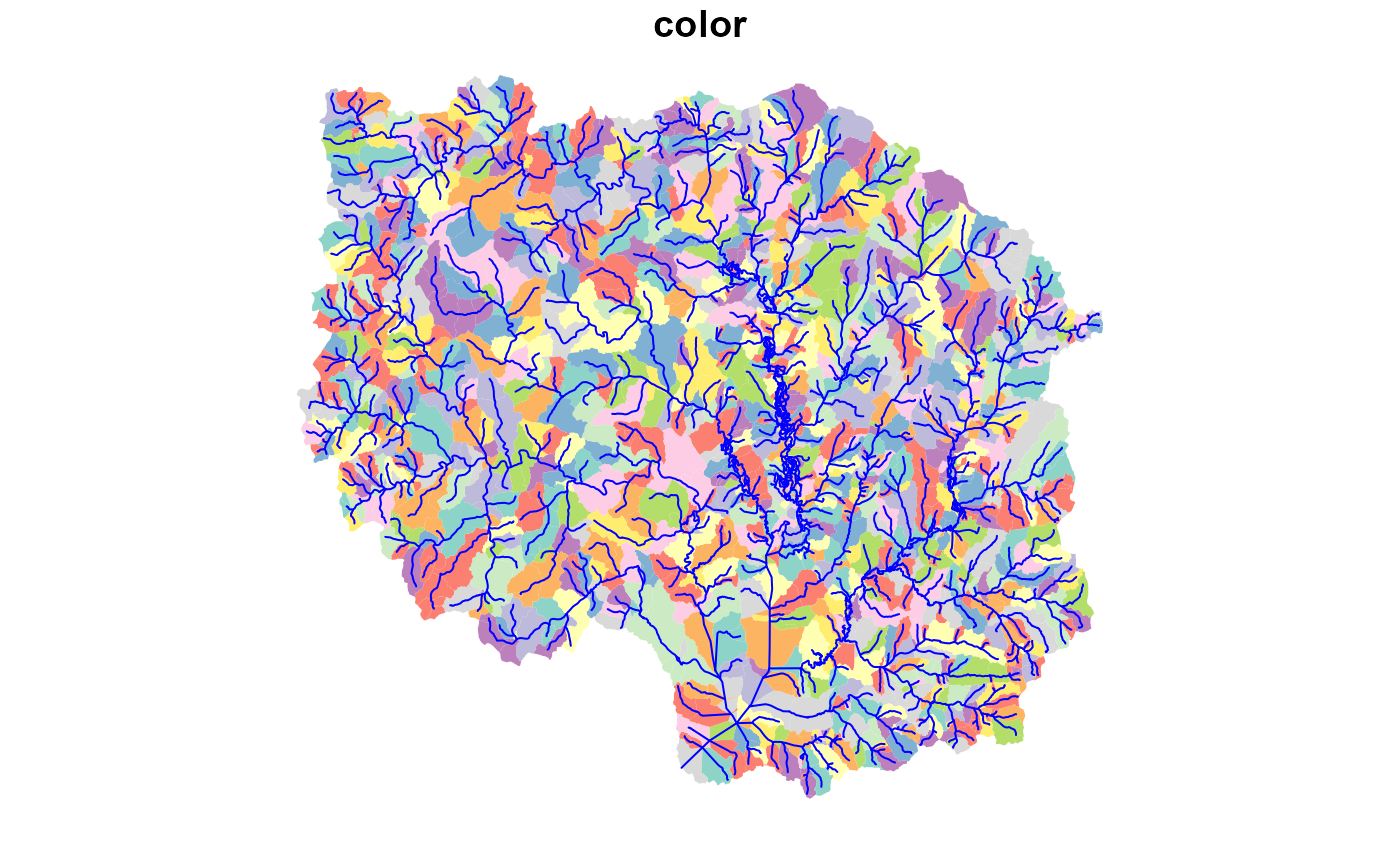
plot(hr_catchment["color"], border = NA, reset = FALSE)
plot(sf::st_geometry(hr_flowline), col = "blue", add = TRUE)
pfaf <- get_pfaf(fl, max_level = 4)
#> Warning: get_pfaf is deprecated, please use hydroloom
hr_catchment <- left_join(hr_data$NHDPlusCatchment, pfaf, by = c("FEATUREID" = "ID"))
colors <- data.frame(pf_level_4 = unique(hr_catchment$pf_level_4),
color = sample(terrain.colors(length(unique(hr_catchment$pf_level_4)))),
stringsAsFactors = FALSE)
hr_catchment <- left_join(hr_catchment, colors, by = "pf_level_4")
plot(hr_catchment["color"], border = NA, reset = FALSE)
plot(sf::st_geometry(hr_flowline), col = "blue", add = TRUE)
 source(system.file("extdata", "walker_data.R", package = "nhdplusTools"))
fl <- select(walker_flowline, COMID, AreaSqKM) %>%
right_join(prepare_nhdplus(walker_flowline, 0, 0,
purge_non_dendritic = FALSE, warn = FALSE),
by = "COMID") %>%
sf::st_sf() %>%
select(ID = COMID, toID = toCOMID, area = AreaSqKM)
fl$nameID = ""
fl$totda <- calculate_total_drainage_area(sf::st_set_geometry(fl, NULL))
#> Warning: calculate_total_drainage_area() is deprecated. Please switch to hydroloom equivalent.
#> Dendritic routing will be applied. Diversions are assumed to have 0 flow fraction.
fl <- left_join(fl, get_levelpaths(rename(sf::st_set_geometry(fl, NULL),
weight = totda)), by = "ID")
#> Warning: get_levelpaths is deprecated in favor of add_levelpaths in the hydroloom package.
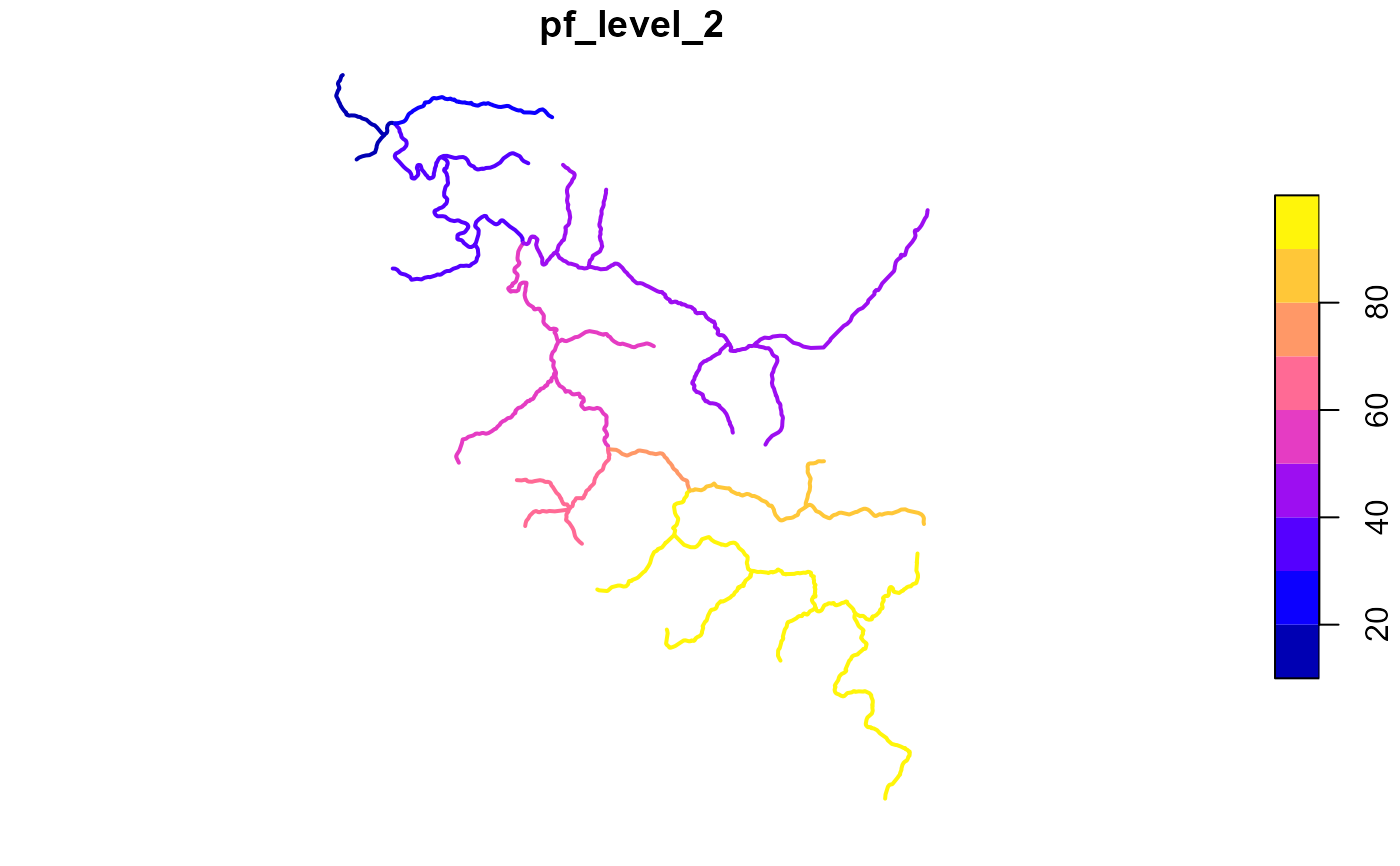
pfaf <- get_pfaf(fl, max_level = 2)
#> Warning: get_pfaf is deprecated, please use hydroloom
fl <- left_join(fl, pfaf, by = "ID")
plot(fl["pf_level_2"], lwd = 2)
source(system.file("extdata", "walker_data.R", package = "nhdplusTools"))
fl <- select(walker_flowline, COMID, AreaSqKM) %>%
right_join(prepare_nhdplus(walker_flowline, 0, 0,
purge_non_dendritic = FALSE, warn = FALSE),
by = "COMID") %>%
sf::st_sf() %>%
select(ID = COMID, toID = toCOMID, area = AreaSqKM)
fl$nameID = ""
fl$totda <- calculate_total_drainage_area(sf::st_set_geometry(fl, NULL))
#> Warning: calculate_total_drainage_area() is deprecated. Please switch to hydroloom equivalent.
#> Dendritic routing will be applied. Diversions are assumed to have 0 flow fraction.
fl <- left_join(fl, get_levelpaths(rename(sf::st_set_geometry(fl, NULL),
weight = totda)), by = "ID")
#> Warning: get_levelpaths is deprecated in favor of add_levelpaths in the hydroloom package.
pfaf <- get_pfaf(fl, max_level = 2)
#> Warning: get_pfaf is deprecated, please use hydroloom
fl <- left_join(fl, pfaf, by = "ID")
plot(fl["pf_level_2"], lwd = 2)
 # }
# }