Note
Go to the end to download the full example code.
3D Point Cloud class
from geobipy import Point
from os.path import join
import numpy as np
import matplotlib.pyplot as plt
import h5py
nPoints = 200
Create a quick test example using random points $z=x(1-x)cos(4pi x)sin(4pi y^{2})^{2}$
x = -np.abs((2.0 * np.random.rand(nPoints)) - 1.0)
y = -np.abs((2.0 * np.random.rand(nPoints)) - 1.0)
z = x * (1.0 - x) * np.cos(np.pi * x) * np.sin(np.pi * y)
PC3D = Point(x=x, y=y, z=z)
Append pointclouds together
x = np.abs((2.0 * np.random.rand(nPoints)) - 1.0)
y = np.abs((2.0 * np.random.rand(nPoints)) - 1.0)
z = x * (1.0 - x) * np.cos(np.pi * x) * np.sin(np.pi * y)
Other_PC = Point(x=x, y=y, z=z)
PC3D.append(Other_PC)
<geobipy.src.classes.pointcloud.Point.Point object at 0x15d8bd710>
Write a summary of the contents of the point cloud
print(PC3D.summary)
Point
x:
| StatArray
| Name: Easting (m)
| Address:['0x15e618d50']
| Shape: (400,)
| Values: [-0.71137954 -0.3793497 -0.30956525 ... 0.14065745 0.05106131
| 0.88015259]
| Min: -0.9976593326444143
| Max: 0.9920098541714664
| has_posterior: False
y:
| StatArray
| Name: Northing (m)
| Address:['0x15e61a750']
| Shape: (400,)
| Values: [-0.37888241 -0.21170869 -0.82494396 ... 0.71680437 0.78865183
| 0.60356845]
| Min: -0.9966527811460972
| Max: 0.9990626592727034
| has_posterior: False
z:
| StatArray
| Name: Height (m)
| Address:['0x15e61b8d0']
| Shape: (400,)
| Values: [-0.6966743 0.11948867 0.11933325 ... 0.08488321 0.02947636
| -0.09294835]
| Min: -1.9925193760132849
| Max: 0.22842565578586502
| has_posterior: False
elevation:
| StatArray
| Name: Elevation (m)
| Address:['0x15e61bad0']
| Shape: (400,)
| Values: [0. 0. 0. ... 0. 0. 0.]
| Min: 0.0
| Max: 0.0
| has_posterior: False
Get a single location from the point as a 3x1 vector
Point = PC3D[50]
# Print the point to the screen
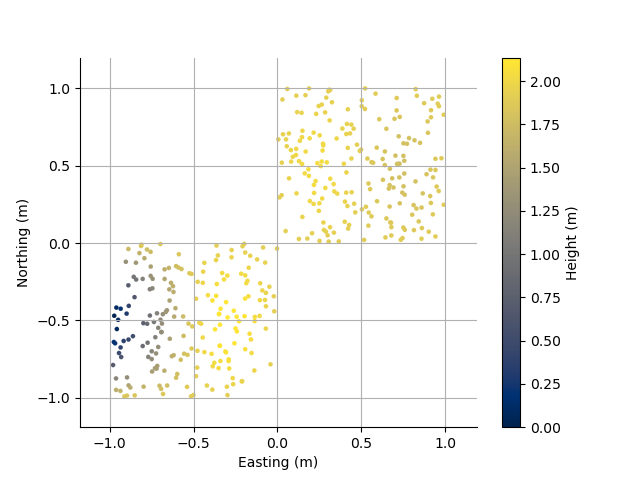
Plot the locations with Height as colour
plt.figure()
PC3D.scatter2D(edgecolor='k')
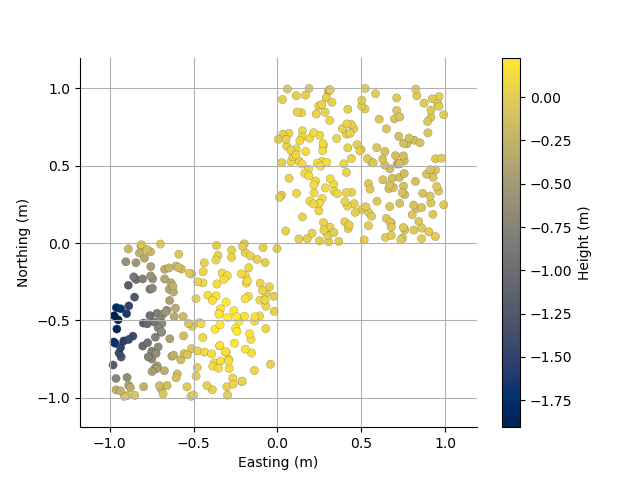
(<Axes: xlabel='Easting (m)', ylabel='Northing (m)'>, <matplotlib.collections.PathCollection object at 0x15e37bb60>, <matplotlib.colorbar.Colorbar object at 0x15e378050>)
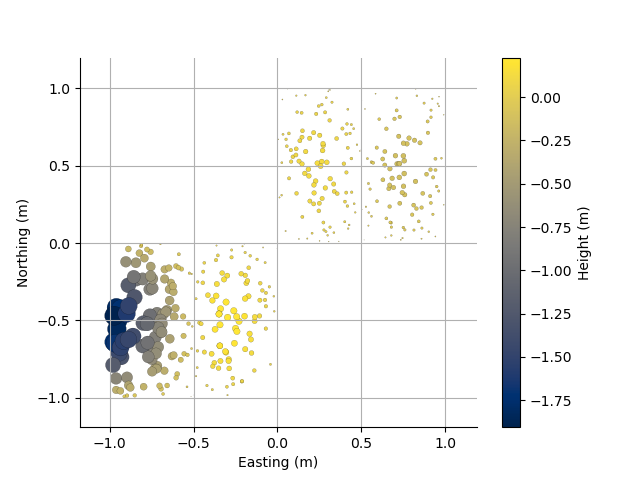
Plotting routines take matplotlib arguments for customization
For example, plotting the size of the points according to the absolute value of height
plt.figure()
ax = PC3D.scatter2D(s=100*np.abs(PC3D.z), edgecolor='k')
Interpolate the points to a 2D rectilinear mesh
mesh, dum = PC3D.interpolate(0.01, 0.01, values=PC3D.z, method='sibson', mask=0.03)
# We can save that mesh to VTK
PC3D.to_vtk('pc3d.vtk')
mesh.to_vtk('interpolated_pc3d.vtk')
Grid the points using a triangulated CloughTocher, or minimum curvature interpolation
plt.figure()
plt.subplot(331)
PC3D.map(dx=0.01, dy=0.01, method='ct')
plt.subplot(332)
PC3D.map(dx=0.01, dy=0.01, method='mc')
plt.subplot(333)
PC3D.map(dx=0.01, dy=0.01, method='sibson')
plt.subplot(334)
PC3D.map(dx=0.01, dy=0.01, method='ct', mask=0.03)
plt.subplot(335)
PC3D.map(dx=0.01, dy=0.01, method='mc', mask=0.3)
plt.subplot(336)
PC3D.map(dx=0.01, dy=0.01, method='sibson', mask=0.03)
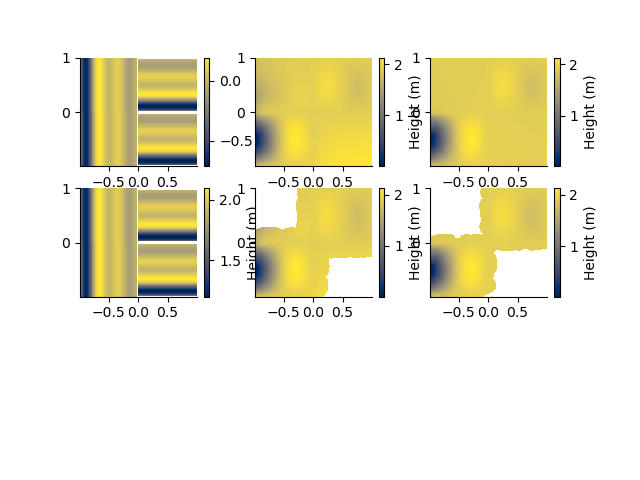
surface [WARNING]: 2 unusable points were supplied; these will be ignored.
surface [WARNING]: You should have pre-processed the data with block-mean, -median, or -mode.
surface [WARNING]: Check that previous processing steps write results with enough decimals.
surface [WARNING]: Possibly some data were half-way between nodes and subject to IEEE 754 rounding.
surface [WARNING]: 2 unusable points were supplied; these will be ignored.
surface [WARNING]: You should have pre-processed the data with block-mean, -median, or -mode.
surface [WARNING]: Check that previous processing steps write results with enough decimals.
surface [WARNING]: Possibly some data were half-way between nodes and subject to IEEE 754 rounding.
(<Axes: >, <matplotlib.collections.QuadMesh object at 0x15e482c00>, <matplotlib.colorbar.Colorbar object at 0x15e898d40>)
For lots of points, these surfaces can look noisy. Using a block filter will help
PCsub = PC3D.block_median(0.05, 0.05)
plt.subplot(337)
PCsub.map(dx=0.01, dy=0.01, method='ct', mask=0.03)
plt.subplot(338)
PCsub.map(dx=0.01, dy=0.01, method='mc', mask=0.03)
plt.subplot(339)
PCsub.map(dx=0.01, dy=0.01, method='sibson', mask=0.03)
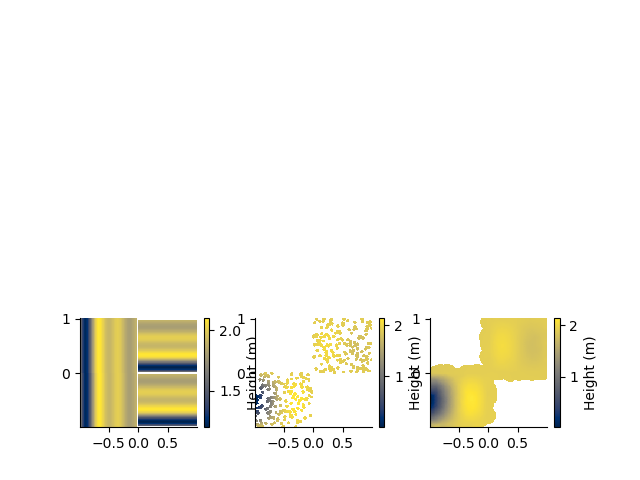
surface [WARNING]: 2 unusable points were supplied; these will be ignored.
surface [WARNING]: You should have pre-processed the data with block-mean, -median, or -mode.
surface [WARNING]: Check that previous processing steps write results with enough decimals.
surface [WARNING]: Possibly some data were half-way between nodes and subject to IEEE 754 rounding.
(<Axes: >, <matplotlib.collections.QuadMesh object at 0x15d6e0c20>, <matplotlib.colorbar.Colorbar object at 0x15daf4a70>)
We can perform spatial searches on the 3D point cloud
PC3D.set_kdtree(ndim=2)
p = PC3D.nearest((0.0,0.0), k=200, p=2, radius=0.3)
.nearest returns the distances and indices into the point cloud of the nearest points. We can then obtain those points as another point cloud
# pNear = PC3D[p[1]]
# plt.figure()
# ax1 = plt.subplot(1,2,1)
# pNear.scatter2D()
# plt.plot(0.0, 0.0, 'x')
# plt.subplot(1,2,2, sharex=ax1, sharey=ax1)
# ax, sc, cb = PC3D.scatter2D(edgecolor='k')
# searchRadius = plt.Circle((0.0, 0.0), 0.3, color='b', fill=False)
# ax.add_artist(searchRadius)
# plt.plot(0.0, 0.0, 'x')
Read in the xyz co-ordinates in columns 2,3,4 from a file. Skip 1 header line.
dataFolder = "..//..//supplementary//Data//"
PC3D.read_csv(filename=dataFolder + 'Resolve1.txt')
<geobipy.src.classes.pointcloud.Point.Point object at 0x15e61e4d0>
plt.figure()
f = PC3D.scatter2D(s=10)
with h5py.File('test.h5', 'w') as f:
PC3D.createHdf(f, 'test')
PC3D.writeHdf(f, 'test')
with h5py.File('test.h5', 'r') as f:
PC3D1 = Point.fromHdf(f['test'])
with h5py.File('test.h5', 'r') as f:
point = Point.fromHdf(f['test'], index=0)
plt.show()
Total running time of the script: (0 minutes 2.708 seconds)