Note
Go to the end to download the full example code.
Skytem dataset
from geobipy import plotting as cP
from os.path import join
import matplotlib.pyplot as plt
import numpy as np
from geobipy import StatArray
from geobipy import TdemData
import h5py
Reading in the Data
dataFolder = "..//..//supplementary//data//"
# The data file name
dataFiles=dataFolder + 'skytem_saline_clay.csv'
# dataFiles = dataFolder + 'Skytem.csv'
# The EM system file name
systemFiles=[dataFolder + 'SkytemHM.stm', dataFolder + 'SkytemLM.stm']
from pathlib import Path
for f in systemFiles[:1]:
txt = Path(f).read_text()
print(txt)
System Begin
Name = SkyTemHighMoment-ElkHills
Type = Time Domain
Transmitter Begin
NumberOfTurns = 1
PeakCurrent = 1
LoopArea = 1
BaseFrequency = 30.0
WaveformDigitisingFrequency = 491520
WaveFormCurrent Begin
-4.00E-03 0.00E+00
-3.91E-03 3.17E-01
-3.81E-03 6.30E-01
-3.72E-03 8.79E-01
-3.68E-03 9.61E-01
-2.30E-03 9.74E-01
-1.01E-03 9.88E-01
0.00E+00 1.00E+00
3.25E-06 9.91E-01
1.00E-04 7.02E-01
2.02E-04 3.78E-01
2.82E-04 1.16E-01
3.08E-04 2.79E-02
3.13E-04 1.21E-02
3.15E-04 6.61E-03
3.17E-04 3.03E-03
3.19E-04 0.00E+00
0.012666667 0.00E+00
WaveFormCurrent End
Transmitter End
Receiver Begin
NumberOfWindows = 26
WindowWeightingScheme = AreaUnderCurve
WindowTimes Begin
3.796E-04 3.872E-04
3.876E-04 3.972E-04
3.976E-04 4.102E-04
4.106E-04 4.262E-04
4.266E-04 4.462E-04
4.466E-04 4.712E-04
4.716E-04 5.022E-04
5.026E-04 5.422E-04
5.426E-04 5.932E-04
5.936E-04 6.562E-04
6.566E-04 7.372E-04
7.376E-04 8.382E-04
8.386E-04 9.652E-04
9.656E-04 1.126E-03
1.127E-03 1.328E-03
1.329E-03 1.583E-03
1.584E-03 1.905E-03
1.906E-03 2.311E-03
2.312E-03 2.822E-03
2.823E-03 3.468E-03
3.469E-03 4.260E-03
4.261E-03 5.228E-03
5.229E-03 6.413E-03
6.414E-03 7.865E-03
7.866E-03 9.641E-03
9.642E-03 1.182E-02
WindowTimes End
LowPassFilter Begin
CutOffFrequency = 300000 210000
Order = 1 2
LowPassFilter End
Receiver End
ForwardModelling Begin
//TX loop area is was 340.82 m^2 -> r = sqrt(340.82/pi)
ModellingLoopRadius = 10.416
OutputType = dB/dt
XOutputScaling = 0
YOutputScaling = 0
ZOutputScaling = 1
SecondaryFieldNormalisation = none
FrequenciesPerDecade = 5
NumberOfAbsiccaInHankelTransformEvaluation = 21
ForwardModelling End
System End
Read in the data from file
TD = TdemData.read_csv(dataFiles, systemFiles)
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
Plot the locations of the data points
plt.figure(1, figsize=(8,6))
_ = TD.scatter2D()
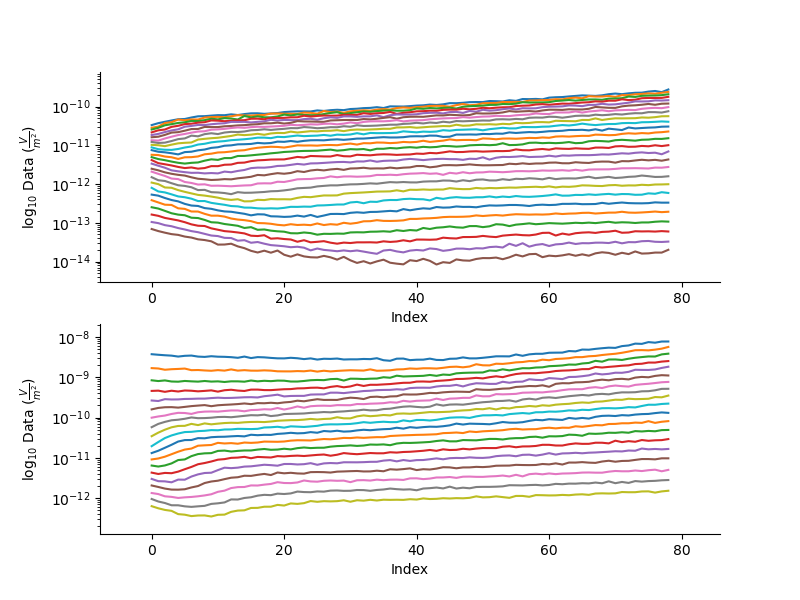
Plot all the data along the specified line
plt.figure(2, figsize=(8,6))
_ = TD.plotLine(0.0, log=10)
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
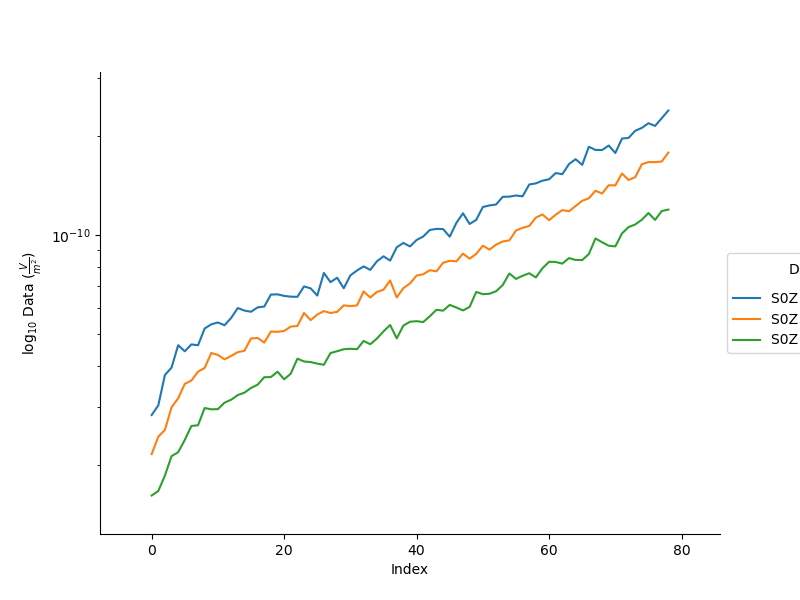
Or, plot specific channels in the data
plt.figure(3, figsize=(8,6))
_ = TD.plot_data(system=0, channels=[1, 3, 5], log=10)
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
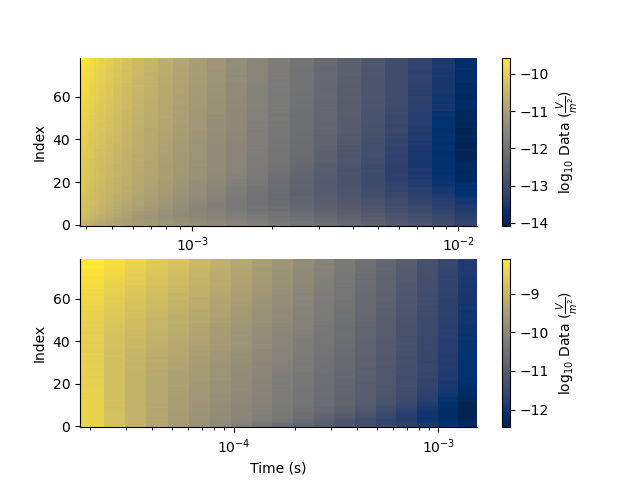
plt.figure(4)
plt.subplot(211)
_ = TD.pcolor(system=0, xscale='log', log=10)
plt.subplot(212)
_ = TD.pcolor(system=1, xscale='log', log=10)
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
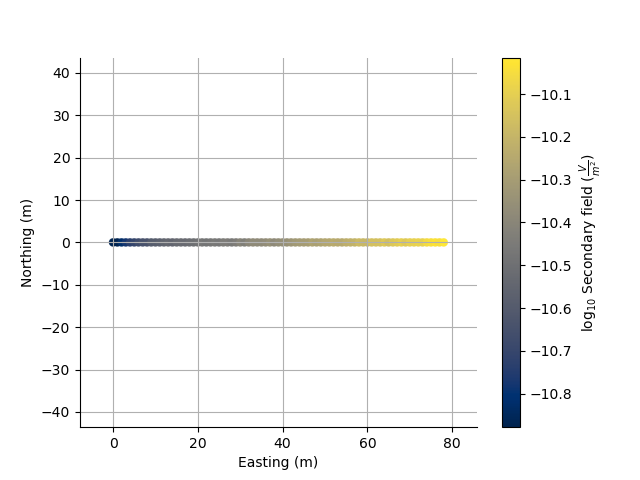
plt.figure(5)
ax = TD.scatter2D(c=TD.secondary_field[:, TD.channel_index(system=0, channel=6)], log=10)
plt.axis('equal')
# with h5py.File('tdem.h5', 'w') as f:
# TD.createHdf(f, 'tdem')
# TD.writeHdf(f, 'tdem')
# with h5py.File('tdem.h5', 'r') as f:
# TD3 = TdemData.fromHdf(f['tdem'])
# with h5py.File('tdem.h5', 'r') as f:
# tdp = TdemData.fromHdf(f['tdem'], index=0)
# #%%
# # Obtain a line from the data set
# # +++++++++++++++++++++++++++++++
# line = TD.line(0.0)
# #%%
# plt.figure(6)
# _ = line.scatter2D(c=line.secondary_field[:, line.channel_index(system=0, channel=6)], log=10)
# #%%
# plt.figure(7)
# _ = line.plot(xAxis='index', log=10)
# Prepare the dataset so that we can read a point at a time.
Dataset = TdemData._initialize_sequential_reading(dataFiles, systemFiles)
# Get a datapoint from the file.
DataPoint = Dataset._read_record()
plt.show()
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
self.n_components=1, self.nTimes=array([26, 19])
File Format for time domain data
Here we describe the file format for time domain data.
The data columns are read in according to the column names in the first line
In this description, the column name or its alternatives are given followed by what the name represents Optional columns are also described.
Required columns
- line
Line number for the data point
- fid
Unique identification number of the data point
- x or northing or n
Northing co-ordinate of the data point, (m)
- y or easting or e
Easting co-ordinate of the data point, (m)
- z or alt
Altitude of the transmitter coil above ground level (m)
- elevation
Elevation of the ground at the data point (m)
- txrx_dx
Distance in x between transmitter and reciever (m)
- txrx_dy
Distance in y between transmitter and reciever (m)
- txrx_dz
Distance in z between transmitter and reciever (m)
- Tx_Pitch
Pitch of the transmitter loop
- Tx_Roll
Roll of the transmitter loop
- Tx_Yaw
Yaw of the transmitter loop
- Rx_Pitch
Pitch of the receiver loop
- Rx_Roll
Roll of the receiver loop
- Rx_Yaw
Yaw of the receiver loop
- Off_time[0] Off_time[1] … Off_time[last] - with the number and square brackets
The measurements for each time gate specified in the accompanying system file under Receiver Window Times The total number of off_time columns should equal the sum of the receiver windows in all system files.
Optional columns
- Off_time_Error[0] Off_time_Error[1] … Off_time_Error[last]
Estimates of standard deviation for each off time measurement
Example Header
Line fid easting northing elevation height txrx_dx txrx_dy txrx_dz TxPitch TxRoll TxYaw RxPitch RxRoll RxYaw Off[0] Off[1]
File Format for a time domain system
Please see Page 13 of Ross Brodie’s instructions
We use GA-AEM for our airborne time domain forward modeller.
Example system files are contained in the supplementary folder in this repository
Total running time of the script: (0 minutes 5.187 seconds)