08: Modflow-setup demonstration
Modflow-setup is a Python package for automating construction of MODFLOW models from grid-independent source data including shapefiles, rasters, and other MODFLOW models that are geo-located. Input data and model construction options are summarized in a single configuration file in the YAML format. Source data are read from their native formats and mapped to a regular finite difference grid specified in the configuration file. An external array-based Flopy model instance with the desired packages is created from the sampled source data and configuration settings. Since every modeling problem is different, the Flopy model instance can be further modified in the same script, using Flopy or other Python packages. MODFLOW input can then be written from the flopy model instance.
Example problem
Here we use Modflow-setup to make an inset model within the Pleasant Lake example model illustrated in exercise 03: Loading and visualizing groundwater models.
The inset model will be the same as the “parent” model, except
the layer top and bottom elevations, head observations and Lake and SFR Package boundary conditions will be re-discretized horizontally on a finer grid.
The Streamflow Routing (SFR) Packages is constructed from a NHDPlus v2 dataset
The Lake package is created from a polygon feature, bathymetry raster, stage-area-volume file and climate data from PRISM.
Lake package observations are set up automatically (with an output file for each lake)
Layer 1 in the parent model is subdivided into layers 1 and 2 in the inset model
Head observations are (re)specified from csv files
Array inputs to the RCHa, NPF, STO Packages will be resampled (interpolated) from the parent model to the inset model grid.
Pumping (Well Package) wells represented in the parent model will be mapped to the nearest cell center in the inset model.
As we’ll see below, Modflow-setup does all of this automatically, based on input specified in a configuration file.
*.py file) that runs from start to finish. To get started with Modflow-setup on your project, see the 10 Minutes to Modflow-setup guide, which includes template configuration files and model build scripts that you can download.Part 1) The configuration file
The configuration file is the primary vehicle of user input to Modflow-setup. The goal is to have a human and machine-readable document that concisely summarizes the data sources and construction methods/options for the basic model structure prior to parameter estimation. Input is specified in the YAML format, which is analogous to a Python dictionary but with nesting indicated by indention instead of curly braces.
Contents of data/pleasant.yml:
# input for MODFLOW 6 simulation
simulation:
sim_name: 'pleasant-inset'
version: 'mf6'
sim_ws: 'pleasant-inset/'
# input for MODFLOW 6 model
model:
simulation: 'pleasant-inset'
modelname: 'pleasant-inset'
options:
print_input: True
save_flows: True
newton: True
newton_under_relaxation: True
# packages to build
# (any packages not listed or commented out will not be built,
# event if they have an input block below)
packages: ['dis',
'ic',
'npf',
'oc',
'sto',
# Note: with no recharge block below and default_source_data=True,
# recharge is regridded from parent model
'rch',
'sfr',
'lak',
'obs',
'wel',
'ims',
'chd'
]
# Regional model to extract boundary conditions, property arrays, and pumping data from
parent:
# arguments to flopy.modflow.Modflow.load for parent model
namefile: 'pleasant.nam'
model_ws: 'pleasant-lake/'
version: 'mf6'
# modflow-setup options
# default_source_data: True, packages and variables that are omitted
# will be pulled from the parent model
default_source_data: True
copy_stress_periods: 'all'
inset_layer_mapping: # mapping between inset and parent model layers
0: 0 # inset: parent (inset layers 1 and 0 copied from parent layer 0)
1: 0
2: 1
3: 2
4: 3
start_date_time: '2012-01-01'
length_units: 'meters'
time_units: 'days'
SpatialReference:
xoff: 552400
yoff: 387200
epsg: 3070
# parameters for setting up the horizontal configuration of the grid
setup_grid:
xoff: 553200 # lower left x-coordinate
yoff: 387800 # lower left y-coordinate
rotation: 0.
dxy: 50 # uniform cell spacing, in CRS units of meters
crs: 3070 # Coordinate reference system (Wisconsin Transverse Mercator)
# Structured Discretization Package
dis:
options:
length_units: 'meters'
dimensions:
nlay: 5
nrow: 100
ncol: 108
source_data:
top:
# DEM file; path relative to setup script
filename: 'pleasant-lake/source_data/rasters/dem40m.tif'
elevation_units: 'meters'
botm:
filenames: # preprocessed layer surfaces
1: 'pleasant-lake/source_data/rasters/botm0.tif'
2: 'pleasant-lake/source_data/rasters/botm1.tif'
3: 'pleasant-lake/source_data/rasters/botm2.tif'
4: 'pleasant-lake/source_data/rasters/botm3.tif'
# Temporal Discretization Package
tdis:
options:
time_units: 'days'
start_date_time: '2012-01-01'
perioddata:
# time discretization info can be specified directly under the perioddata key
# or in groups of stress periods that are discretized in a similar way
group 1: # initial steady-state period (steady specified under sto package)
#perlen: 1 # Specify perlen as an int or list of lengths in model units,
# or perlen=None and 3 of start_date, end_date, nper and/or freq."
nper: 1
nstp: 1
tsmult: 1
# "steady" can be entered here;
# otherwise the global entry specified in the STO package is used as the default
steady: True
# oc_saverecord: can also be specified by group here;
# otherwise the global entry specified in the oc package is used as the default
group 2: # monthly stress periods
# can be specified by group,
# otherwise start_date_time for the model (under tdis: options) will be used.
start_date_time: '2012-01-01'
# model ends at midnight on this date (2012-12-31 would be the last day simulated)
end_date_time: '2013-01-01'
freq: '1MS' # see arguments to pandas.date_range
nstp: 1
tsmult: 1.5
steady: False
# Initial Conditions Package
ic:
source_data:
strt:
from_parent: # starting heads sampled from parent model
binaryfile: 'pleasant-lake/pleasant.hds'
stress_period: 0
# Node Property Flow Package
npf:
options:
save_flows: True
griddata:
icelltype: 1 # variable sat. thickness in all layers
# with parent: default_source_data: True,
# unspecified variables such as "k" and "k33" are resampled from parent model
# Storage Package
sto:
options:
save_flows: True
griddata:
iconvert: 1 # convertible layers
# with parent: default_source_data: True,
# unspecified variables such as "sy" and "ss" are resampled from parent model
# Well Package
wel:
options:
print_input: True
print_flows: True
save_flows: True
# with parent: default_source_data: True,
# unspecified well fluxes are resampled from parent model
# Streamflow Routing Package
# SFR input is created using SFRmaker (https://github.com/doi-usgs/sfrmaker)
sfr:
options:
save_flows: True
source_data:
flowlines:
# path to NHDPlus version 2 dataset
nhdplus_paths: ['pleasant-lake/source_data/shps']
# if a DEM is included, streambed top elevations will be sampled
# from the minimum DEM values within a 100 meter buffer around each stream reach
dem:
filename: 'pleasant-lake/source_data/rasters/dem40m.tif'
elevation_units: 'meters'
# SFR observations can be automatically setup from a CSV file
# of x, y locations and fluxes
observations: # see sfrmaker.observations.add_observations for arguments
filename: 'pleasant-lake/source_data/tables/gages.csv'
obstype: 'downstream-flow' # modflow-6 observation type
x_location_column: 'x'
y_location_column: 'y'
obsname_column: 'site_no'
sfrmaker_options:
# the sfrmaker_options: block can include arguments
# to the Lines.to_sfr method in SFRmaker
# or other options such as set_streambed_top_elevations_from_dem
# or to_riv (see SFRmaker and Modflow-setup docs)
set_streambed_top_elevations_from_dem: True
# Lake Package
lak:
options:
boundnames: True
save_flows: True
surfdep: 0.1
source_data:
# initial lakebed leakance rates
littoral_leakance: 0.045 # for thin zone around lake perimeter; 1/d
profundal_leakance: 0.025 # for interior of lake basin; 1/d
littoral_zone_buffer_width: 40
lakes_shapefile: # polygon shapefile of lake footprints
filename: 'pleasant-lake/source_data/shps/all_lakes.shp'
id_column: 'HYDROID'
include_ids: [600059060] # pleasant lake
# setup lake precipitation and evaporation from PRISM data
climate:
filenames:
600059060: 'pleasant-lake/source_data/PRISM_ppt_tmean_stable_4km_189501_201901_43.9850_-89.5522.csv'
format: 'prism'
period_stats:
# for period 0, use average precip and evap for dates below
0: ['mean', '2012-01-01', '2018-12-31'] # average daily rate for model period for initial steady state
# for subsequent periods,
# average precip and evap to period start/end dates
1: 'mean' # average daily rate or value for each month
# bathymetry file with lake depths to subtract off model top
bathymetry_raster:
filename: 'pleasant-lake/source_data/rasters/pleasant_bathymetry.tif'
length_units: 'meters'
# bathymetry file with stage/area/volume relationship
stage_area_volume_file:
filename: 'pleasant-lake/source_data/tables/area_stage_vol_Pleasant.csv'
length_units: 'meters'
id_column: 'hydroid'
column_mappings:
volume_m3: 'volume'
external_files: False # option to write connectiondata table to external file
# Iterative model solution
ims:
options:
complexity: 'complex'
nonlinear:
outer_dvclose: 1.e-1
outer_maximum: 200
# Observation (OBS) Utility
obs:
source_data:
# Head observations are supplied via csv files
# with x, y locations and observation names
# an observation is generated in each model layer;
# observations at each location can subsequently be processed
# to represent a given screened interval
# for example, using modflow-obs (https://github.com/aleaf/modflow-obs)
filenames: ['pleasant-lake/source_data/tables/nwis_heads_info_file.csv',
'pleasant-lake/source_data/tables/lake_sites.csv',
'pleasant-lake/source_data/tables/wdnr_gw_sites.csv',
'pleasant-lake/source_data/tables/uwsp_heads.csv',
'pleasant-lake/source_data/tables/wgnhs_head_targets.csv'
]
column_mappings:
obsname: ['obsprefix', 'obsnme', 'common_name']
drop_observations: ['10019209_lk' # pleasant lake; monitored via Lake Package observations
]
# Perimeter boundary condition of heads extracted from parent model solution
chd:
perimeter_boundary:
parent_head_file: 'pleasant-lake/pleasant.hds'
Part 2) The model build script
A template model build script that can be further customized for your project is available here.
[1]:
import os
from pathlib import Path
import numpy as np
import pandas as pd
from scipy.interpolate import griddata
import matplotlib.pyplot as plt
from matplotlib import patheffects
import flopy
from mfsetup import MF6model
Define some functions
A function to just make a model grid
Oftentimes at the start of a modeling project, we want to quickly test different grid resolutions and extents before attempting to build the model. We can do this with Modflow-setup by creating a model instance and then running the setup_grid() method. A Flopy-like model grid instance is created from the setup_grid: block in the configuration file.
Wrapping this in a function helps to
keep the
__main__part of the script clean and concise, andalso provides a more robust way to control the current working directory (
cwd). Modflow-setup changes thecwdto the specified model workspace (model_ws:specified above), to more easily work with external (MODFLOW) input files through the Flopy interface.
[2]:
def setup_grid(configuration_file):
"""Just set up (a shapefile of) the model grid.
For trying different grid configurations."""
cwd = os.getcwd()
model = MF6model(cfg=configuration_file)
model.setup_grid()
model.modelgrid.write_shapefile('postproc/shps/grid.shp')
os.chdir(cwd)
return model
A function to build the model using Modflow-setup
[3]:
def setup_model(configuration_file):
"""Set up the whole model."""
cwd = os.getcwd()
model = MF6model.setup_from_yaml(configuration_file)
model.write_input()
os.chdir(cwd)
return model
Specify the model configuration file
Read the model workspace location and model name from the configuration file, so that we only have to specify them there.
[4]:
import yaml
model_configuration_file = Path('data/pleasant.yml')
# Load the model configuration file into a dictionary
with open(model_configuration_file) as src:
cfg = yaml.load(src, Loader=yaml.Loader)
sim_ws = model_configuration_file.parent / cfg['simulation']['sim_ws']
model_name = cfg['model']['modelname']
Just make a model grid
[5]:
%%capture
model = setup_grid('data/pleasant.yml')
[6]:
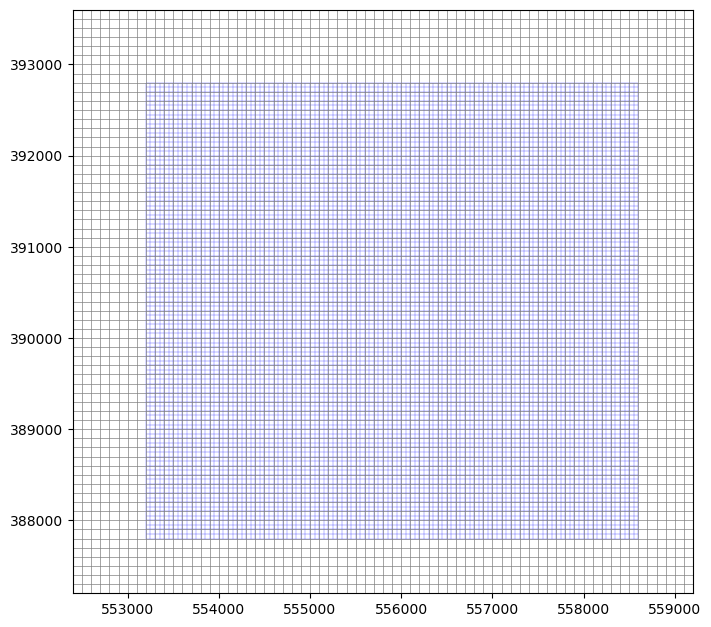
fig, ax = plt.subplots(figsize=(8, 8))
model.modelgrid.plot(ax=ax, zorder=2, ec='b', lw=0.2)
model.parent.modelgrid.plot(ax=ax, lw=0.5)
ax.set_ylim(*model.parent.modelgrid.bounds[1::2])
ax.set_xlim(*model.parent.modelgrid.bounds[::2])
[6]:
(552400.0, 559200.0)
[7]:
parent_model = model.parent
parent_model.modelgrid.write_shapefile(sim_ws / 'postproc/shps/regional_model_grid.shp')
creating shapely Polygons of grid cells...
finished in 0.06s
writing data/pleasant-inset/postproc/shps/regional_model_grid.shp... Done
Build the whole model
[8]:
%%capture
model = setup_model('data/pleasant.yml')
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[9], line 1
----> 1 model = setup_model('data/pleasant.yml')
Cell In[3], line 4, in setup_model(configuration_file)
2 """Set up the whole model."""
3 cwd = os.getcwd()
----> 4 model = MF6model.setup_from_yaml(configuration_file)
5 model.write_input()
6 os.chdir(cwd)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/mfmodel.py:1819, in MFsetupMixin.setup_from_yaml(cls, yamlfile, verbose)
1806 """Make a model from scratch, using information in a yamlfile.
1807
1808 Parameters
(...) 1816 m : model instance
1817 """
1818 cfg = cls.load_cfg(yamlfile, verbose=verbose)
-> 1819 return cls.setup_from_cfg(cfg, verbose=verbose)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/mfmodel.py:1855, in MFsetupMixin.setup_from_cfg(cls, cfg, verbose)
1852 m.setup_solver()
1854 # set up all of the packages specified in the config file
-> 1855 m.setup_packages(reset_existing=False)
1857 # LGR inset model(s)
1858 if m.inset is not None:
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/mfmodel.py:1784, in MFsetupMixin.setup_packages(self, reset_existing)
1782 # avoid multiple package instances for now, except for obs
1783 if self.version != 'mf6' or pkg == 'obs' or not hasattr(self, pkg):
-> 1784 package_setup(**self.cfg[pkg], **self.cfg[pkg]['mfsetup_options'])
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/mf6model.py:543, in MF6model.setup_dis(self, **kwargs)
538 self._setup_array(package, 'top', datatype='array2d',
539 resample_method='linear',
540 write_fmt='%.2f')
542 # make the botm array
--> 543 self._setup_array(package, 'botm', datatype='array3d',
544 resample_method='linear',
545 write_fmt='%.2f')
547 # set number of layers to length of the created bottom array
548 # this needs to be set prior to setting up the idomain,
549 # otherwise idomain may have wrong number of layers
550 self.cfg['dis']['dimensions']['nlay'] = len(self.cfg['dis']['griddata']['botm'])
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/mfmodel.py:1251, in MFsetupMixin._setup_array(self, package, var, vmin, vmax, source_model, source_package, **kwargs)
1248 def _setup_array(self, package, var, vmin=-1e30, vmax=1e30,
1249 source_model=None, source_package=None,
1250 **kwargs):
-> 1251 return setup_array(self, package, var, vmin=vmin, vmax=vmax,
1252 source_model=source_model, source_package=source_package,
1253 **kwargs)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/sourcedata.py:1424, in setup_array(model, package, var, data, vmin, vmax, datatype, source_model, source_package, write_fmt, write_nodata, **kwargs)
1421 # copy the preexisting top file
1422 shutil.copy(model.cfg['intermediate_data']['top'][0],
1423 original_top_file)
-> 1424 top = model.load_array(original_top_file)
1425 if model.version == 'mf6':
1426 # reset the model top to the lake bottom
1427 top[bathy != 0] -= bathy[bathy != 0]
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/mfmodel.py:560, in MFsetupMixin.load_array(self, filename)
554 arrays.append(load_array(f,
555 shape=(self.nrow, self.ncol),
556 nodata=self._nodata_value
557 )
558 )
559 return np.array(arrays)
--> 560 return load_array(filename, shape=(self.nrow, self.ncol))
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/mfsetup/fileio.py:127, in load_array(filename, shape, nodata)
124 print(txt, end=', ')
125 # arr = np.loadtxt
126 # pd.read_csv is >3x faster than np.load_txt
--> 127 arr = pd.read_csv(f, delim_whitespace=True, header=None).values
128 if shape2d is not None:
129 if arr.shape != shape2d:
TypeError: read_csv() got an unexpected keyword argument 'delim_whitespace'
a MF6model instance (subclass of flopy.mf6.ModflowGwf) is returned
[9]:
model
/home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-inset
[9]:
Pleasant Lake test case version 0.1.post562.dev0+g97bd77b
Parent model: /home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-lake//pleasant
100 row(s), 108 column(s)
delr: [50.00...50.00] undefined
delc: [50.00...50.00] undefined
CRS: EPSG:3070
length units: meters
xll: 553200.0; yll: 387800.0; rotation: 0.0
Bounds: (np.float64(553200.0), np.float64(558600.0), np.float64(387800.0), np.float64(392800.0))
Packages:
13 period(s):
per start_datetime end_datetime perlen steady nstp
0 2012-01-01 2012-01-01 1.0 True 1
1 2012-01-01 2012-02-01 31.0 False 1
2 2012-02-01 2012-03-01 29.0 False 1
...
12 2012-12-01 2013-01-01 31.0 False 1
information from the configuration file is stored in an attached cfg dictionary
[10]:
model.cfg.keys()
[10]:
dict_keys(['metadata', 'simulation', 'model', 'parent', 'postprocessing', 'setup_grid', 'dis', 'tdis', 'ic', 'npf', 'sto', 'rch', 'sfr', 'high_k_lakes', 'lak', 'mvr', 'chd', 'drn', 'ghb', 'riv', 'wel', 'oc', 'obs', 'ims', 'mfsetup_options', 'filename', 'maw', 'external_files', 'intermediate_data', 'grid'])
the cfg dictionary contains both information from the configuration file, and MODFLOW input (such as external text file arrays) that was developed from the original source data. Internally in Modflow-setup, MODFLOW input in cfg is fed to the various Flopy object constructors.
[11]:
model.cfg['dis']
[11]:
defaultdict(dict,
{'remake_top': True,
'options': {'length_units': 'meters'},
'dimensions': {'nlay': 5, 'nrow': 100, 'ncol': 108},
'griddata': {},
'top_filename_fmt': 'top.dat',
'botm_filename_fmt': 'botm_{:03d}.dat',
'idomain_filename_fmt': 'idomain_{:03d}.dat',
'minimum_layer_thickness': 1,
'drop_thin_cells': True,
'source_data': {'top': {'filename': '/home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-lake/source_data/rasters/dem40m.tif',
'elevation_units': 'meters'},
'botm': {'filenames': {1: '/home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-lake/source_data/rasters/botm0.tif',
2: '/home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-lake/source_data/rasters/botm1.tif',
3: '/home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-lake/source_data/rasters/botm2.tif',
4: '/home/runner/work/python-for-hydrology/python-for-hydrology/docs/source/notebooks/part1_flopy/data/pleasant-lake/source_data/rasters/botm3.tif'}}},
'nlay': 4})
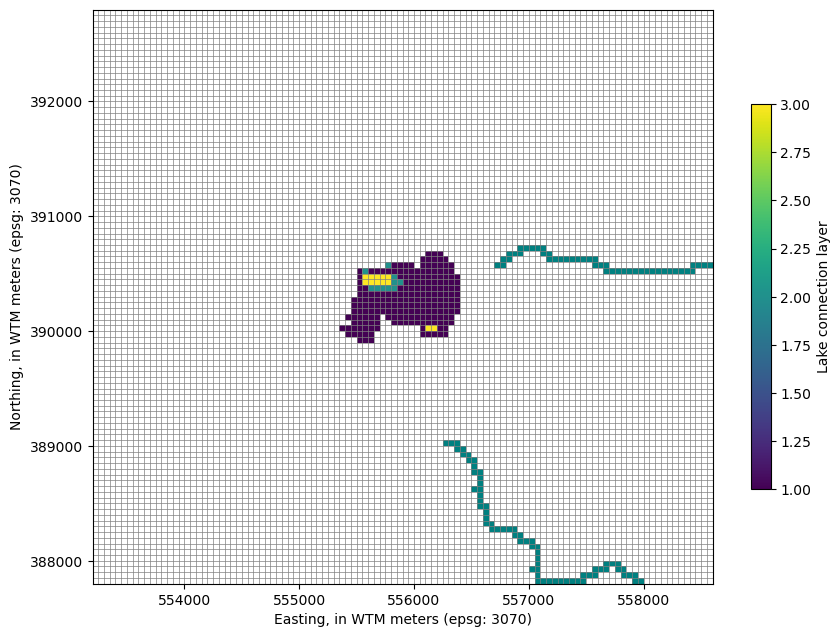
Plot the model grid with Lake Package connections by layer
[12]:
# Get the parent model grid extent
# to set plot limits later
modelgrid = model.modelgrid
l, r, b, t = modelgrid.extent
# Make the plot
fig, ax = plt.subplots(figsize=(10, 10))
mv = flopy.plot.PlotMapView(model=model, ax=ax)
mv.plot_bc("SFR", plotAll=True)
lcp = mv.plot_grid(lw=0.5, ax=ax)
# Get the lake connections
vconn = model.lak.connectiondata.array[model.lak.connectiondata.array['claktype'] == 'vertical']
k, i, j = zip(*vconn['cellid'])
lakeconnections = np.zeros((modelgrid.nrow, modelgrid.ncol))
lakeconnections[i, j] = np.array(k) + 1
lakeconnections = np.ma.masked_array(lakeconnections, mask=lakeconnections == 0)
qmi = mv.plot_array(lakeconnections)
# re-limit the plot to the parent model extent
ax.set_ylim(b, t)
ax.set_xlim(l, r)
ax.set_aspect(1)
plt.colorbar(qmi, shrink=0.5, label='Lake connection layer')
ax.set_xlabel('Easting, in WTM meters (epsg: 3070)')
ax.set_ylabel('Northing, in WTM meters (epsg: 3070)')
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[12], line 9
7 fig, ax = plt.subplots(figsize=(10, 10))
8 mv = flopy.plot.PlotMapView(model=model, ax=ax)
----> 9 mv.plot_bc("SFR", plotAll=True)
10 lcp = mv.plot_grid(lw=0.5, ax=ax)
12 # Get the lake connections
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/plot/map.py:494, in PlotMapView.plot_bc(self, name, package, kper, color, plotAll, boundname, **kwargs)
491 raise Exception("Cannot find package to plot")
493 # trap for mf6 'cellid' vs mf2005 'k', 'i', 'j' convention
--> 494 if isinstance(p, list) or p.parent.version == "mf6":
495 if not isinstance(p, list):
496 p = [p]
AttributeError: 'NoneType' object has no attribute 'parent'
Run the model
we already wrote the input above in the
setup_model()function (while the working directory was set to the model workspace)
Note: Running the model through Flopy (as below) requires specification of the MODFLOW executable. In Flopy, the executable is specified via the exe_name argument to the simulation constructor for MODFLOW 6, or model constructor for previous MODFLOW versions. Similarly, in Modflow-setup, the exe_name is specified in the simulation: or model: block of the configuration file.
This example assumes that a MODFLOW 6 executable with the name “mf6” either resides in the model workspace, or is included in the system path.
[13]:
model.simulation.run_simulation()
FloPy is using the following executable to run the model: ../../../../../../../../../.local/bin/mf6
ERROR REPORT:
1. mf6: mfsim.nam is not present in working directory.
[13]:
(False, [])
Plot the results
Read the output and post-process the 3D head results into a 2D water table
[14]:
import flopy.utils.binaryfile as bf
from flopy.utils.postprocessing import get_water_table
headfile_object = bf.HeadFile(sim_ws / 'pleasant-inset.hds')
# read the head results for the last stress period
kper = 12
heads = headfile_object.get_data(kstpkper=(0, kper))
# Get the water table elevation from the 3D head results
water_table = get_water_table(heads)
# put in the lake level (not included in head output)
lake_results = pd.read_csv(sim_ws / 'lake1.obs.csv')
stage = lake_results['STAGE'][kper]
cnd = pd.DataFrame(model.lak.connectiondata.array)
k, i, j = zip(*cnd['cellid'])
water_table[i, j] = stage
# add the SFR stage as well
sfr_stage = model.sfr.output.stage().get_data()[0, 0, :]
# get the SFR cell i, j locations
# by unpacking the cellid tuples in the packagedata
sfr_k, sfr_i, sfr_j = zip(*model.sfr.packagedata.array['cellid'])
water_table[sfr_i, sfr_j] = sfr_stage
# get the cell budget using the .output attribute
# (instead of the binaryfile utility directly)
cbc = model.output.budget()
lake_fluxes = cbc.get_data(text='lak', full3D=True)[0].sum(axis=0)
sfr_fluxes = cbc.get_data(text='sfr', full3D=True)[0]
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
Cell In[14], line 4
1 import flopy.utils.binaryfile as bf
2 from flopy.utils.postprocessing import get_water_table
----> 4 headfile_object = bf.HeadFile(sim_ws / 'pleasant-inset.hds')
6 # read the head results for the last stress period
7 kper = 12
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/utils/binaryfile/__init__.py:504, in HeadFile.__init__(self, filename, text, precision, verbose, **kwargs)
502 self.text = text.encode()
503 if precision == "auto":
--> 504 precision = get_headfile_precision(filename)
505 if precision == "unknown":
506 s = f"Error. Precision could not be determined for {filename}"
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/utils/binaryfile/__init__.py:249, in get_headfile_precision(filename)
246 result = "unknown"
248 # Open file, and check filesize to ensure this is not an empty file
--> 249 f = open(filename, "rb")
250 f.seek(0, 2)
251 totalbytes = f.tell()
FileNotFoundError: [Errno 2] No such file or directory: 'data/pleasant-inset/pleasant-inset.hds'
Make the plot
show “leakage” results for the Lake and SFR packages (neg. values indicate groundwater discharge to surface water)
[15]:
levels=np.arange(280, 315, 2)
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(model, ax=ax)
ctr = pmv.contour_array(water_table, levels=levels,
linewidths=.5, colors='b')
labels = pmv.ax.clabel(ctr, inline=True, fontsize=8, inline_spacing=1)
vmin, vmax = -100, 100
im = pmv.plot_array(lake_fluxes, cmap='coolwarm', vmin=vmin, vmax=vmax)
im = pmv.plot_array(sfr_fluxes.sum(axis=0), cmap='coolwarm', vmin=vmin, vmax=vmax)
cb = fig.colorbar(im, shrink=0.7, label='Leakage, in m$^3$/day')
ax.set_ylabel("Northing, WTM meters")
ax.set_xlabel("Easting, WTM meters")
ax.set_aspect(1)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[15], line 5
3 fig, ax = plt.subplots(figsize=(8, 8))
4 pmv = flopy.plot.PlotMapView(model, ax=ax)
----> 5 ctr = pmv.contour_array(water_table, levels=levels,
6 linewidths=.5, colors='b')
7 labels = pmv.ax.clabel(ctr, inline=True, fontsize=8, inline_spacing=1)
8 vmin, vmax = -100, 100
NameError: name 'water_table' is not defined
What if we want to further customize the model?
Add a high-capacity pumping well at a specified location, using the Multi-Aquifer Well (MAW) Package, which isn’t yet supported by Modflow-setup
[16]:
well_x, well_y = 556000, 389500
# get the row, column location from the coordinates
well_i, well_j = model.modelgrid.intersect(well_x, well_y)
well_i, well_j
[16]:
(np.int64(65), np.int64(55))
Set up the MAW Package
[17]:
#wellno radius bottom strt condeqn ngwfnodes boundname
packagedata = [[0, 0.25, 274, 296., 'mean', 4, 'new_well']]
#wellno icon k i j scrn_top scrn_botm hk_skin radius_skin
connectiondata = [
[0, 0, 0, well_i, well_j, 299, 274, 100.0, 0.7],
[0, 1, 1, well_i, well_j, 299, 274, 100.0, 0.7],
[0, 2, 2, well_i, well_j, 299, 274, 100.0, 0.7],
[0, 3, 3, well_i, well_j, 299, 274, 100.0, 0.7]
]
perioddata_input = [[0, "rate", -2000*1440/264.17]]
maw = flopy.mf6.ModflowGwfmaw(
model,
boundnames=True,
save_flows=True,
packagedata=packagedata,
connectiondata=connectiondata,
perioddata=perioddata_input,
pname='maw'
)
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatalist.py:396, in MFList._set_data(self, data, autofill, check_data, preserve_record)
395 # store data
--> 396 self._get_storage_obj().set_data(
397 data,
398 autofill=autofill,
399 check_data=check_data,
400 preserve_record=preserve_record,
401 )
402 except Exception as ex:
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatastorage.py:923, in DataStorage.set_data(self, data, layer, multiplier, key, autofill, check_data, preserve_record)
919 if (
920 self.data_structure_type == DataStructureType.recarray
921 or self.data_structure_type == DataStructureType.scalar
922 ):
--> 923 self._set_list(
924 data,
925 layer,
926 multiplier,
927 key,
928 autofill,
929 check_data,
930 preserve_record,
931 )
932 else:
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatastorage.py:995, in DataStorage._set_list(self, data, layer, multiplier, key, autofill, check_data, preserve_record)
994 return
--> 995 self.store_internal(
996 data,
997 layer,
998 False,
999 multiplier,
1000 key=key,
1001 autofill=autofill,
1002 check_data=check_data,
1003 preserve_record=preserve_record,
1004 )
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatastorage.py:1374, in DataStorage.store_internal(self, data, layer, const, multiplier, key, autofill, print_format, check_data, preserve_record)
1373 if data is not None:
-> 1374 new_data = self._build_recarray(data, key, autofill)
1375 self.layer_storage.first_item().internal_data = (
1376 new_data
1377 )
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatastorage.py:1598, in DataStorage._build_recarray(self, data, key, autofill)
1597 if self._simulation_data.verify_data:
-> 1598 self._verify_list(new_data)
1599 return new_data
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatastorage.py:2277, in DataStorage._verify_list(self, data)
2272 cellid_size = self.data_dimensions.get_cellid_size(
2273 self._recarray_type_list[index][0]
2274 )
2275 if (
2276 cellid_size != 1
-> 2277 and len(data_line[index]) != cellid_size
2278 and isinstance(data_line[index], int)
2279 ):
2280 message = (
2281 'Cellid "{}" contains {} integer(s). '
2282 "Expected a cellid containing {} "
(...) 2289 )
2290 )
TypeError: object of type 'int' has no len()
During handling of the above exception, another exception occurred:
MFDataException Traceback (most recent call last)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatalist.py:85, in MFList.__init__(self, sim_data, model_or_sim, structure, data, enable, path, dimensions, package, block)
84 try:
---> 85 self.set_data(data, True)
86 except Exception as ex:
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatalist.py:571, in MFList.set_data(self, data, autofill, check_data)
551 """Sets the contents of the data to "data". Data can have the
552 following formats:
553 1) recarray - recarray containing the datalist
(...) 569
570 """
--> 571 self._set_data(data, autofill, check_data=check_data)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatalist.py:404, in MFList._set_data(self, data, autofill, check_data, preserve_record)
403 type_, value_, traceback_ = sys.exc_info()
--> 404 raise MFDataException(
405 self.structure.get_model(),
406 self.structure.get_package(),
407 self._path,
408 "setting data",
409 self.structure.name,
410 inspect.stack()[0][3],
411 type_,
412 value_,
413 traceback_,
414 None,
415 self._simulation_data.debug,
416 ex,
417 )
418 if check_data and self._simulation_data.verify_data:
419 # verify cellids
MFDataException: An error occurred in data element "connectiondata" model "gwf6" package "maw". The error occurred while setting data in the "_set_data" method.
During handling of the above exception, another exception occurred:
MFDataException Traceback (most recent call last)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/mfpackage.py:622, in MFBlock.add_dataset(self, dataset_struct, data, var_path)
621 try:
--> 622 self.datasets[var_path[-1]] = self.data_factory(
623 self._simulation_data,
624 self._model_or_sim,
625 dataset_struct,
626 True,
627 var_path,
628 self._dimensions,
629 data,
630 self._container_package,
631 )
632 except MFDataException as mfde:
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/mfpackage.py:494, in MFBlock.data_factory(self, sim_data, model_or_sim, structure, enable, path, dimensions, data, package)
493 else:
--> 494 return mfdatalist.MFList(
495 sim_data,
496 model_or_sim,
497 structure,
498 data,
499 enable,
500 path,
501 dimensions,
502 package,
503 self,
504 )
505 elif data_type == mfstructure.DataType.list_transient:
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/data/mfdatalist.py:88, in MFList.__init__(self, sim_data, model_or_sim, structure, data, enable, path, dimensions, package, block)
87 type_, value_, traceback_ = sys.exc_info()
---> 88 raise MFDataException(
89 structure.get_model(),
90 structure.get_package(),
91 path,
92 "setting data",
93 structure.name,
94 inspect.stack()[0][3],
95 type_,
96 value_,
97 traceback_,
98 None,
99 sim_data.debug,
100 ex,
101 )
MFDataException: An error occurred in data element "connectiondata" model "gwf6" package "maw". The error occurred while setting data in the "__init__" method.
During handling of the above exception, another exception occurred:
MFDataException Traceback (most recent call last)
Cell In[17], line 12
4 connectiondata = [
5 [0, 0, 0, well_i, well_j, 299, 274, 100.0, 0.7],
6 [0, 1, 1, well_i, well_j, 299, 274, 100.0, 0.7],
7 [0, 2, 2, well_i, well_j, 299, 274, 100.0, 0.7],
8 [0, 3, 3, well_i, well_j, 299, 274, 100.0, 0.7]
9 ]
10 perioddata_input = [[0, "rate", -2000*1440/264.17]]
---> 12 maw = flopy.mf6.ModflowGwfmaw(
13 model,
14 boundnames=True,
15 save_flows=True,
16 packagedata=packagedata,
17 connectiondata=connectiondata,
18 perioddata=perioddata_input,
19 pname='maw'
20 )
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/modflow/mfgwfmaw.py:1142, in ModflowGwfmaw.__init__(self, model, loading_package, auxiliary, boundnames, print_input, print_head, print_flows, save_flows, head_filerecord, budget_filerecord, budgetcsv_filerecord, no_well_storage, flow_correction, flowing_wells, shutdown_theta, shutdown_kappa, mfrcsv_filerecord, timeseries, observations, mover, nmawwells, packagedata, connectiondata, perioddata, filename, pname, **kwargs)
1140 self.nmawwells = self.build_mfdata("nmawwells", nmawwells)
1141 self.packagedata = self.build_mfdata("packagedata", packagedata)
-> 1142 self.connectiondata = self.build_mfdata("connectiondata", connectiondata)
1143 self.perioddata = self.build_mfdata("perioddata", perioddata)
1145 self._init_complete = True
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/mfpackage.py:2803, in MFPackage.build_mfdata(self, var_name, data)
2801 dataset_struct = block.data_structures[var_name]
2802 var_path = self.path + (key, var_name)
-> 2803 ds = self.blocks[block.name].add_dataset(
2804 dataset_struct, data, var_path
2805 )
2806 self._data_list.append(ds)
2807 return ds
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/flopy/mf6/mfpackage.py:633, in MFBlock.add_dataset(self, dataset_struct, data, var_path)
622 self.datasets[var_path[-1]] = self.data_factory(
623 self._simulation_data,
624 self._model_or_sim,
(...) 630 self._container_package,
631 )
632 except MFDataException as mfde:
--> 633 raise MFDataException(
634 mfdata_except=mfde,
635 model=self._container_package.model_name,
636 package=self._container_package._get_pname(),
637 message="Error occurred while adding"
638 ' dataset "{}" to block '
639 '"{}"'.format(dataset_struct.name, self.structure.name),
640 )
642 self._simulation_data.mfdata[var_path] = self.datasets[var_path[-1]]
643 dtype = dataset_struct.get_datatype()
MFDataException: An error occurred in data element "connectiondata" model "pleasant-inset" package "maw". The error occurred while setting data in the "__init__" method.
Additional Information:
(1) Error occurred while adding dataset "connectiondata" to block "connectiondata"
Write the MAW Package input file;
Refresh the Name file to include the MAW Package
[18]:
model.maw.write()
# refresh the name file with the added package
model.name_file.write()
INFORMATION: nmawwells in ('gwf6', 'maw', 'dimensions') changed to 1 based on size of packagedata
[19]:
model.simulation.run_simulation()
FloPy is using the following executable to run the model: ../../../../../../../../../.local/bin/mf6
ERROR REPORT:
1. mf6: mfsim.nam is not present in working directory.
[19]:
(False, [])
Plot the simulated drawdown
[20]:
heads2 = headfile_object.get_data(kstpkper=(0, kper))
water_table2 = get_water_table(heads2)
lake_results = pd.read_csv(sim_ws / 'lake1.obs.csv')
stage = lake_results['STAGE'][kper]
cnd = pd.DataFrame(model.lak.connectiondata.array)
k, i, j = zip(*cnd['cellid'])
water_table2[i, j] = stage
sfr_stage = model.sfr.output.stage().get_data()[0, 0, :]
sfr_k, sfr_i, sfr_j = zip(*model.sfr.packagedata.array['cellid'])
water_table2[sfr_i, sfr_j] = sfr_stage
drawdown = water_table - water_table2
# get the cell budget using the .output attribute
# (instead of the binaryfile utility directly)
cbc = model.output.budget()
lake_fluxes = cbc.get_data(text='lak', full3D=True)[0].sum(axis=0)
sfr_fluxes = cbc.get_data(text='sfr', full3D=True)[0]
levels=np.arange(1, 10 , 1)
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(model, ax=ax)
ctr = pmv.contour_array(drawdown, levels=levels,
linewidths=.5, colors='b')
labels = pmv.ax.clabel(ctr, inline=True, fontsize=8, inline_spacing=1)
vmin, vmax = -100, 100
im = pmv.plot_array(lake_fluxes, cmap='coolwarm', vmin=vmin, vmax=vmax)
im = pmv.plot_array(sfr_fluxes.sum(axis=0), cmap='coolwarm', vmin=vmin, vmax=vmax)
cb = fig.colorbar(im, shrink=0.7, label='Leakage, in m$^3$/day')
ax.set_ylabel("Northing, WTM meters")
ax.set_xlabel("Easting, WTM meters")
ax.set_aspect(1)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[20], line 1
----> 1 heads2 = headfile_object.get_data(kstpkper=(0, kper))
4 water_table2 = get_water_table(heads2)
5 lake_results = pd.read_csv(sim_ws / 'lake1.obs.csv')
NameError: name 'headfile_object' is not defined