06: FloPy class project: Voronoi grid version
[1]:
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
import geopandas as gpd
import flopy
from flopy.discretization import VertexGrid
from flopy.utils.triangle import Triangle as Triangle
from flopy.utils.voronoi import VoronoiGrid
from flopy.utils.gridintersect import GridIntersect
[2]:
from project_grid_functions import densify_polyline, circle_function
[3]:
model_ws = "../temp/voronoi/"
grid_ws = f"{model_ws}grid/"
Path(grid_ws).mkdir(parents=True, exist_ok=True)
Path('figures').mkdir(parents=True, exist_ok=True)
output_folder = Path('../figures')
output_folder.mkdir(parents=True, exist_ok=True)
Load a few raster files
[4]:
bottom = flopy.utils.Raster.load("../data_project/aquifer_bottom.asc")
top = flopy.utils.Raster.load("../data_project/aquifer_top.asc")
kaq = flopy.utils.Raster.load("../data_project/aquifer_k.asc")
Load a few shapefiles with geopandas
[5]:
river = gpd.read_file("../data_project/Flowline_river.shp")
inactive = gpd.read_file("../data_project/inactive_area.shp")
active = gpd.read_file("../data_project/active_area.shp")
wells = gpd.read_file("../data_project/pumping_well_locations.shp")
Plot the shapefiles with geopandas
[6]:
ax = river.plot(color="cyan")
active.plot(ax=ax, color="blue")
inactive.plot(ax=ax, color="white")
wells.plot(ax=ax, color="red", markersize=8);
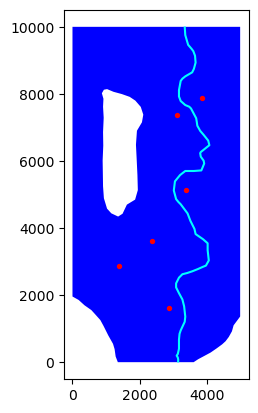
Make a voronoi model grid
Densify the river vertices.
[7]:
river_densify = densify_polyline(river.geometry[0], 200)
river_densify.shape, np.unique(river_densify, axis=0).shape
[7]:
((96, 2), (96, 2))
Modify the end points of river_densify to constrain the river to be within the bounds of the active domain
[8]:
river_densify[0, 1] = 9999.99
river_densify[-1, 1] = 0.1
Run triangle to generate the nodes
[9]:
maximum_area = 500.0 * 500.0
tri = Triangle(
maximum_area=maximum_area, angle=30, nodes=river_densify, model_ws=grid_ws
)
tri.add_polygon(active.geometry[0])
tri.add_polygon(inactive.geometry[0])
tri.add_hole((1500, 6000))
for idx, point in enumerate(wells.centroid):
center = (point.x, point.y)
tri.add_polygon(circle_function(center=center, radius=100))
tri.add_region(center, attribute=idx, maximum_area=maximum_area)
tri.build(verbose=False)
[10]:
tri.plot()
[10]:
<matplotlib.collections.LineCollection at 0x161981190>
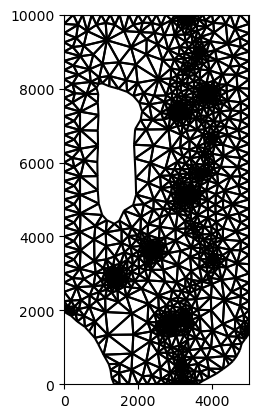
[11]:
vor = VoronoiGrid(tri)
gridprops = vor.get_gridprops_vertexgrid()
idomain = np.ones((1, vor.ncpl), dtype=int)
base_grid = VertexGrid(**gridprops, nlay=1, idomain=idomain)
[12]:
base_grid.plot()
river.plot(color="cyan", ax=plt.gca(), linewidth=0.5)
wells.plot(ax=plt.gca(), color="red", markersize=8, zorder=100);
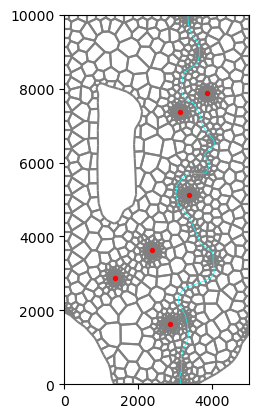
Define the number of layers and some simple shapes
[13]:
nlay = 3
shape2d, shape3d = (base_grid.ncpl), (nlay, base_grid.ncpl)
xlen, ylen = 5000.0, 10000.0
Intersect the modelgrid with the shapefiles
Create an intersection object
[14]:
ix = GridIntersect(base_grid, method="vertex", rtree=True)
Intersect inactive and active shapefiles with the modelgrid
After all of the intersection operations, take a look at the data contained in the returned objects
[15]:
bedrock = ix.intersect(inactive.geometry[0])
active_cells = ix.intersect(active.geometry[0])
[16]:
active_cells[:4], active_cells.dtype
[16]:
(rec.array([(0, <POLYGON ((5000 7875, 4837.765 7875, 4851.886 8125, 5000 8125, 5000 8000, 50...>, 38793.62962818),
(1, <POLYGON ((5000 7750, 5000 7625, 4927.811 7625, 4805.222 7837.36, 4837.765 7...>, 35065.60370048),
(2, <POLYGON ((5000 7500, 5000 7375, 4925.376 7375, 4855.031 7498.923, 4927.811 ...>, 27295.68972587),
(3, <POLYGON ((5000 7250, 5000 7125, 4927.811 7125, 4855.031 7251.077, 4925.376 ...>, 27295.68972587)],
dtype=[('cellids', 'O'), ('ixshapes', 'O'), ('areas', '<f8')]),
dtype((numpy.record, [('cellids', 'O'), ('ixshapes', 'O'), ('areas', '<f8')])))
Intersect well shapefile with the modelgrid
[17]:
well_cells = []
for g in wells.geometry:
v = ix.intersect(g)
well_cells += v["cellids"].tolist()
[18]:
well_cells
[18]:
[1315, 1421, 1435, 1286, 1420, 1434]
Intersect river shapefile with the modelgrid
[19]:
river_cells = ix.intersect(river.geometry[0])
[20]:
river_cells[:4], river_cells.dtype
[20]:
(rec.array([(367, <LINESTRING (3358.332 9833.087, 3361.701 9782.55)>, 50.6490377 ),
(368, <LINESTRING (3361.701 9782.55, 3363.599 9754.072, 3382.073 9691.263)>, 94.01139569),
(369, <LINESTRING (3401.559 9625.008, 3434.568 9512.777)>, 116.98441041),
(370, <LINESTRING (3434.568 9512.777, 3449.104 9463.355, 3500.407 9403.502)>, 130.34665378)],
dtype=[('cellids', 'O'), ('ixshapes', 'O'), ('lengths', '<f8')]),
dtype((numpy.record, [('cellids', 'O'), ('ixshapes', 'O'), ('lengths', '<f8')])))
Intersect constant head line with the modelgrid
Use a line with two points to defined the location of the constant head cells. The line verticase are [(1250, 0.1), (4250, 0.1)].
[21]:
constant_cells = ix.intersect(
[(1250, 0.1), (4250, 0.1)], shapetype="linestring"
)
[22]:
constant_cells[:4], constant_cells.dtype
[22]:
(rec.array([(38, <LINESTRING (3556.433 0.1, 3613.02 0.1)>, 56.58677601),
(39, <LINESTRING (3437.5 0.1, 3556.433 0.1)>, 118.93334885),
(40, <LINESTRING (3234.375 0.1, 3281.25 0.1)>, 46.875 ),
(41, <LINESTRING (2937.5 0.1, 3031.25 0.1)>, 93.75 )],
dtype=[('cellids', 'O'), ('ixshapes', 'O'), ('lengths', '<f8')]),
dtype((numpy.record, [('cellids', 'O'), ('ixshapes', 'O'), ('lengths', '<f8')])))
Resample the raster data to the modelgrid
Use the resample_to_grid() method on each raster object.
[23]:
rtop = top.resample_to_grid(
base_grid,
band=top.bands[0],
method="linear",
extrapolate_edges=True,
)
rbot = bottom.resample_to_grid(
base_grid,
band=bottom.bands[0],
method="linear",
extrapolate_edges=True,
)
rkaq = (
kaq.resample_to_grid(
base_grid, band=kaq.bands[0], method="linear", extrapolate_edges=True
)
* 86400.0
)
Plot the resampled data
Plot the aquifer top, bottom, and hydraulic conductivity. Also plot the aquifer thickness.
[24]:
mm = flopy.plot.PlotMapView(modelgrid=base_grid)
cb = mm.plot_array(rtop)
mm.plot_grid(lw=0.5, color="0.5")
plt.colorbar(cb, ax=plt.gca());
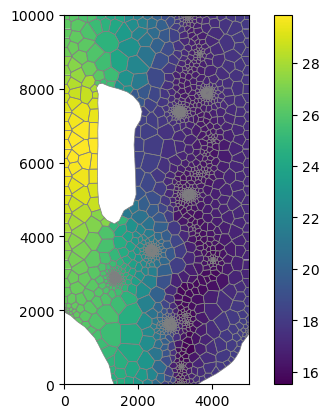
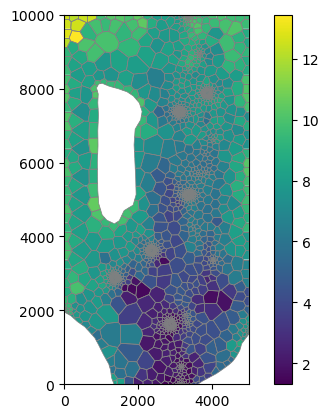
[25]:
mm = flopy.plot.PlotMapView(modelgrid=base_grid)
cb = mm.plot_array(rbot)
mm.plot_grid(lw=0.5, color="0.5")
plt.colorbar(cb, ax=plt.gca());
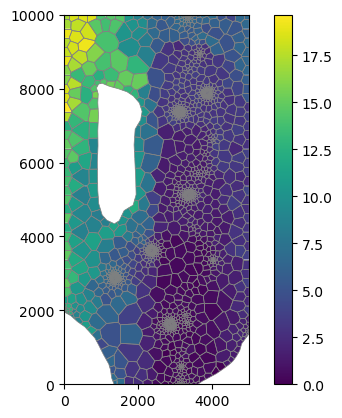
[26]:
mm = flopy.plot.PlotMapView(modelgrid=base_grid)
cb = mm.plot_array(rtop - rbot)
mm.plot_grid(lw=0.5, color="0.5")
plt.colorbar(cb);
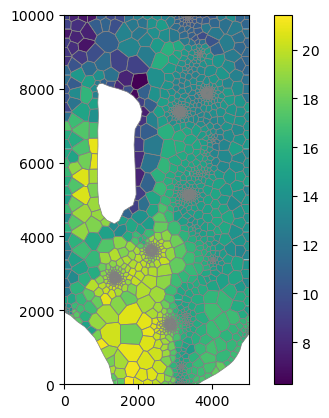
[27]:
mm = flopy.plot.PlotMapView(modelgrid=base_grid)
cb = mm.plot_array(rkaq)
mm.plot_grid(lw=0.5, color="0.5")
plt.colorbar(cb);
Build the model data
Create the bottom of each model layer
Assume that the thickness of each layer at a row, column location is equal.
[28]:
botm = np.zeros(shape3d, dtype=float)
botm[-1, :] = rbot[:]
layer_thickness = (rtop - rbot) / nlay
for k in reversed(range(nlay - 1)):
botm[k] = botm[k + 1] + layer_thickness
Create the idomain array
Use the intersection data from the active and inactive shapefiles to create the idomain array
[29]:
idomain = np.zeros(shape3d, dtype=float)
for node in active_cells["cellids"]:
idomain[:, node] = 1
for node in bedrock["cellids"]:
idomain[:, node] = 0
Build the well package stress period data
The pumping rates are in the
wellsgeopandas dataframePumping rates are in m/sec
The wells are located in model layer 3
[30]:
wells
[30]:
| FID | Q | geometry | |
|---|---|---|---|
| 0 | 0 | -0.00820 | POINT (3875.000 7875.000) |
| 1 | 1 | -0.00410 | POINT (3125.000 7375.000) |
| 2 | 2 | -0.00390 | POINT (3375.000 5125.000) |
| 3 | 3 | -0.00083 | POINT (2375.000 3625.000) |
| 4 | 4 | -0.00072 | POINT (1375.000 2875.000) |
| 5 | 5 | -0.00430 | POINT (2875.000 1625.000) |
[31]:
well_spd = []
for (cellid, q) in zip(well_cells, wells["Q"]):
well_spd.append([2, cellid, q * 86400.0])
well_spd
[31]:
[[2, 1315, -708.48],
[2, 1421, -354.24],
[2, 1435, -336.96],
[2, 1286, -71.712],
[2, 1420, -62.208000000000006],
[2, 1434, -371.52]]
Build the river package stress period data
Calculate the length of the river using the
"lengths"key.The vertical hydraulic conductivity of the river bed sediments is 3.5 m/d.
The thickness of river bottom sediments at the upstream (North) and downstream (South) end of the river is 0.5 and 1.5 meters, respectively.
The river bottom at the upstream and downstream end of the river is 16.5 and 14.5 meters, respectively. The river width at the upstream and downstream end of the river is 5.0 and 10.0 meters, respectively.
The river stage at the upstream and downstream end of the river is 16.6 and 15.5 meters, respectively.
Use the boundname
upstreamfor river cells where the upstream end of the river cell is less than 5000 m from the North end of the model. Use the boundnamedownstreamfor all other river cells.
Use the upstream and downstream values to interpolate the river sediment thickness, bottom, width, and stage for each river cell.
The river cells will be connected to model layer 1. The river bottom, width, and stage values should be calculated at the center of the river reach.
[32]:
river_kv = 3.5
river_thickness_up, river_thickness_down = 0.5, 1.5
river_bot_up, river_bot_down = 16.5, 14.5
river_width_up, river_width_down = 5.0, 10.0
stage_up, stage_down = 16.6, 15.5
[33]:
river_length = river_cells["lengths"].sum()
river_length
[33]:
12419.290359618699
[34]:
river_thickness_slope = (
river_thickness_down - river_thickness_up
) / river_length
[35]:
river_bot_slope = (river_bot_down - river_bot_up) / river_length
[36]:
river_width_slope = (river_width_down - river_width_up) / river_length
[37]:
stage_slope = (stage_down - stage_up) / river_length
[38]:
boundname = "upstream"
total_length = 0.0
river_spd = []
river_top_delta = []
for idx, (cellid, length) in enumerate(
zip(river_cells["cellids"], river_cells["lengths"])
):
if total_length >= 5000.0 and boundname == "upstream":
boundname = "downstream"
dx = 0.5 * length
total_length += dx
river_thickness = river_thickness_up + river_thickness_slope * total_length
river_bot = river_bot_up + river_bot_slope * total_length
river_width = river_width_up + river_width_slope * total_length
river_stage = stage_up + stage_slope * total_length
conductance = river_kv * length * river_width / river_thickness
river_spd.append(
[0, cellid, river_stage, conductance, river_bot, boundname]
)
river_top_delta.append(river_bot - rtop[cellid])
total_length += dx
river_top_delta = np.array(river_top_delta)
river_spd[:2], river_spd[-2:]
[38]:
([[0,
367,
16.597756959542117,
1769.1162067371693,
16.495921744622027,
'upstream'],
[0,
368,
16.5913505356594,
3264.9265367307926,
16.48427370119891,
'upstream']],
[[0,
1882,
15.501184070837642,
218.68249850902666,
14.502152856068442,
'downstream'],
[0,
1944,
15.500384546364442,
202.62120587305378,
14.500699175208076,
'downstream']])
[39]:
river_top_delta.min(), river_top_delta.max(), river_top_delta.mean(), river_top_delta[
-2:
]
[39]:
(-2.23108535744932,
-0.09231567157780418,
-0.8471750038169135,
array([-1.09793806, -1.66048444]))
Define river observations
[40]:
riv_obs = {
"riv_obs.csv": [
("UPSTREAM", "RIV", "UPSTREAM"),
("DOWNSTREAM", "RIV", "DOWNSTREAM"),
],
}
Build SFR datasets
<rno> <cellid(ncelldim)> <rlen> <rwid> <rgrd> <rtp> <rbth> <rhk> <man> <ncon> <ustrf> <ndv> [<aux(naux)>] [<boundname>]
[41]:
nreaches = river_cells.shape[0]
nreaches
[41]:
168
[42]:
boundname = "upstream"
gage1_loc = None
total_length = 0.0
sfr_packagedata = []
sfr_connectivity = []
for idx, (cellid, length) in enumerate(
zip(river_cells["cellids"], river_cells["lengths"])
):
if total_length >= 5000.0 and boundname == "upstream":
boundname = "downstream"
gage1_loc = idx - 1
dx = 0.5 * length
total_length += dx
river_thickness = river_thickness_up + river_thickness_slope * total_length
river_bot = river_bot_up + river_bot_slope * total_length
river_width = river_width_up + river_width_slope * total_length
if idx == 0:
nconn = 1
sfr_connectivity.append((idx, -(idx + 1)))
elif idx == nreaches - 1:
nconn = 1
sfr_connectivity.append((idx, (idx - 1)))
else:
nconn = 2
sfr_connectivity.append((idx, (idx - 1), -(idx + 1)))
sfr_layer = None
for k in range(nlay):
if river_bot - river_thickness > botm[k, cellid]:
sfr_layer = k
if sfr_layer is None:
sfr_cellid = "none"
else:
sfr_cellid = (sfr_layer, cellid)
leakance = river_kv * river_thickness
sfr_packagedata.append(
(
idx,
sfr_cellid,
length,
river_width,
-river_bot_slope,
river_bot,
river_thickness,
river_kv,
0.035,
nconn,
1.0,
0,
boundname,
)
)
total_length += dx
[43]:
sfr_spd = {0: [(0, "inflow", 864000.0)]}
sfr_spd
[43]:
{0: [(0, 'inflow', 864000.0)]}
<rno> <cellid(ncelldim)> <rlen> <rwid> <rgrd> <rtp> <rbth> <rhk> <man> <ncon> <ustrf> <ndv> [<aux(naux)>] [<boundname>]
[44]:
sfr_packagedata[:2], sfr_packagedata[-2:]
[44]:
([(0,
(2, 367),
50.64903769970752,
5.010195638444929,
0.0001610397971290692,
16.495921744622027,
0.5020391276889858,
3.5,
0.035,
1,
1.0,
0,
'upstream'),
(1,
(2, 368),
94.01139569348081,
5.039315747002732,
0.0001610397971290692,
16.48427370119891,
0.5078631494005464,
3.5,
0.035,
2,
1.0,
0,
'upstream')],
[(166,
(2, 1882),
9.370424773718105,
9.994617859828896,
0.0001610397971290692,
14.502152856068442,
1.4989235719657792,
3.5,
0.035,
2,
1.0,
0,
'downstream'),
(167,
(2, 1944),
8.68325992137821,
9.998252061979809,
0.0001610397971290692,
14.500699175208076,
1.4996504123959618,
3.5,
0.035,
1,
1.0,
0,
'downstream')])
[45]:
sfr_connectivity[:2], sfr_connectivity[-2:]
[45]:
([(0, -1), (1, 0, -2)], [(166, 165, -167), (167, 166)])
[46]:
sfr_obs = {
"sfr_obs.csv": [
("UPSTREAM", "SFR", "UPSTREAM"),
("DOWNSTREAM", "SFR", "DOWNSTREAM"),
("GAGE1", "downstream-flow", (gage1_loc,)),
("GAGE2", "ext-outflow", (nreaches - 1,)),
],
}
Define the constant head cells
Assume the constant head cells are located in all three layers and have values equal to the downstream river stage (stage_down). Make sure the constant head stage is greater that the bottom of the layer.
[47]:
chd_spd = []
for node in constant_cells["cellids"]:
for k in range(nlay):
if stage_down > botm[k, node]:
chd_spd.append((k, node, stage_down))
chd_spd[:2], chd_spd[-2:]
[47]:
([(0, 38, 15.5), (1, 38, 15.5)], [(1, 1571, 15.5), (2, 1571, 15.5)])
Define recharge rates
The recharge rate is 0.16000000E-08 m/sec
[48]:
recharge_rate = 0.16000000e-08 * 86400.0
recharge_rate
[48]:
0.00013824
Build the model
Build a steady-state model using the data that you have created. Packages to create:
Simulation
TDIS (1 stress period,
TIME_UNITS='days')IMS (default parameters)
GWF model (
SAVE_FLOWS=True)DISV (
LENGTH_UNITS='meters')IC (
STRT=40.)NPF (Unconfined, same K for all layers, save
SAVE_SPECIFIC_DISCHARGE=True)RCH (array based)
RIV (
BOUNDNAMES=True,addRIVobservations for defined boundnames)CHD
WEL
OC (Save
HEAD ALLandBUDGET ALL)
[49]:
name = "project"
exe_name = "mf6"
exe_name
[49]:
'mf6'
[50]:
sim = flopy.mf6.MFSimulation(
sim_name=name, sim_ws=model_ws, exe_name=str(exe_name)
)
tdis = flopy.mf6.ModflowTdis(sim, time_units="days")
ims = flopy.mf6.ModflowIms(
sim,
linear_acceleration="bicgstab",
outer_maximum=200,
inner_maximum=100,
print_option="all",
)
[51]:
gwf = flopy.mf6.ModflowGwf(
sim,
modelname=name,
save_flows=True,
newtonoptions="NEWTON UNDER_RELAXATION",
)
[52]:
dis = flopy.mf6.ModflowGwfdisv(
gwf,
length_units="meters",
nlay=nlay,
ncpl=base_grid.ncpl,
nvert=base_grid.nvert,
vertices=vor.get_disv_gridprops()["vertices"],
cell2d=vor.get_disv_gridprops()["cell2d"],
top=rtop,
botm=botm,
)
ic = flopy.mf6.ModflowGwfic(gwf, strt=[rtop for k in range(nlay)])
npf = flopy.mf6.ModflowGwfnpf(
gwf,
save_specific_discharge=True,
save_saturation=True,
k=[rkaq, rkaq, rkaq], icelltype=1
)
rch = flopy.mf6.ModflowGwfrcha(gwf, recharge=recharge_rate)
# riv = flopy.mf6.ModflowGwfriv(gwf, boundnames=True, stress_period_data=river_spd)
# riv.obs.initialize(
# filename=f"{name}.riv.obs",
# print_input=True,
# continuous=riv_obs,
# )
sfr = flopy.mf6.ModflowGwfsfr(
gwf,
unit_conversion=86400.0,
boundnames=True,
print_flows=True,
print_stage=True,
nreaches=nreaches,
packagedata=sfr_packagedata,
connectiondata=sfr_connectivity,
perioddata=sfr_spd,
)
sfr.obs.initialize(
filename=f"{name}.sfr.obs",
print_input=True,
continuous=sfr_obs,
)
wel = flopy.mf6.ModflowGwfwel(gwf, stress_period_data=well_spd)
chd = flopy.mf6.ModflowGwfchd(gwf, stress_period_data=chd_spd)
oc = flopy.mf6.ModflowGwfoc(
gwf,
head_filerecord=f"{name}.hds",
budget_filerecord=f"{name}.cbc",
saverecord=[("HEAD", "ALL"), ("BUDGET", "ALL")],
)
Write the model files
[53]:
sim.write_simulation()
writing simulation...
writing simulation name file...
writing simulation tdis package...
writing solution package ims_-1...
writing model project...
writing model name file...
writing package disv...
writing package ic...
writing package npf...
writing package rcha_0...
writing package sfr_0...
writing package obs_0...
writing package wel_0...
INFORMATION: maxbound in ('gwf6', 'wel', 'dimensions') changed to 6 based on size of stress_period_data
writing package chd_0...
INFORMATION: maxbound in ('gwf6', 'chd', 'dimensions') changed to 140 based on size of stress_period_data
writing package oc...
Run the model
[54]:
sim.run_simulation()
FloPy is using the following executable to run the model: ../../../../../../../.local/share/flopy/bin/mf6
MODFLOW 6
U.S. GEOLOGICAL SURVEY MODULAR HYDROLOGIC MODEL
VERSION 6.4.2 06/28/2023
MODFLOW 6 compiled Jul 05 2023 20:29:14 with Intel(R) Fortran Intel(R) 64
Compiler Classic for applications running on Intel(R) 64, Version 2021.7.0
Build 20220726_000000
This software has been approved for release by the U.S. Geological
Survey (USGS). Although the software has been subjected to rigorous
review, the USGS reserves the right to update the software as needed
pursuant to further analysis and review. No warranty, expressed or
implied, is made by the USGS or the U.S. Government as to the
functionality of the software and related material nor shall the
fact of release constitute any such warranty. Furthermore, the
software is released on condition that neither the USGS nor the U.S.
Government shall be held liable for any damages resulting from its
authorized or unauthorized use. Also refer to the USGS Water
Resources Software User Rights Notice for complete use, copyright,
and distribution information.
Run start date and time (yyyy/mm/dd hh:mm:ss): 2024/02/03 11:13:17
Writing simulation list file: mfsim.lst
Using Simulation name file: mfsim.nam
Solving: Stress period: 1 Time step: 1
Run end date and time (yyyy/mm/dd hh:mm:ss): 2024/02/03 11:13:17
Elapsed run time: 0.292 Seconds
WARNING REPORT:
1. OPTIONS BLOCK VARIABLE 'UNIT_CONVERSION' IN FILE 'project.sfr' WAS
DEPRECATED IN VERSION 6.4.2. SETTING UNIT_CONVERSION DIRECTLY.
Normal termination of simulation.
[54]:
(True, [])
Post-process the results
Use gwf.output. method to get the observations.
[55]:
myobs = gwf.sfr.output.obs().get_data()
myobs.dtype
[55]:
dtype([('totim', '<f8'), ('UPSTREAM', '<f8'), ('DOWNSTREAM', '<f8'), ('GAGE1', '<f8'), ('GAGE2', '<f8')])
[56]:
myobs["UPSTREAM"], myobs["DOWNSTREAM"]
[56]:
(array([5516.58121287]), array([-8810.44888484]))
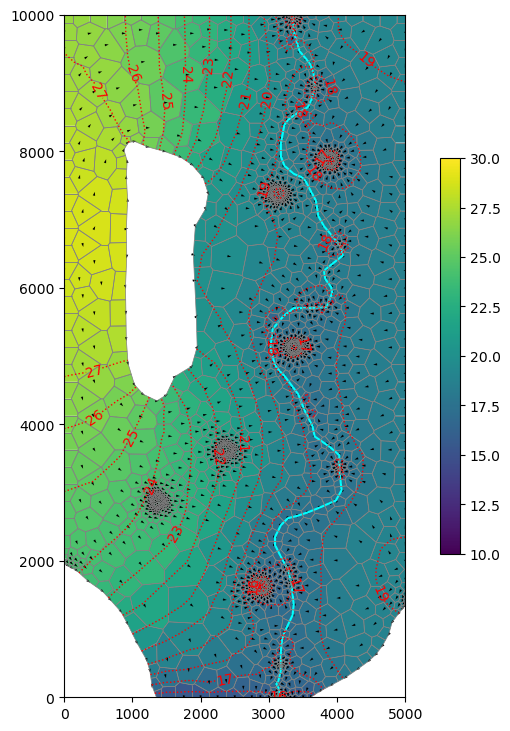
Use gwf.output. method to get the heads and specific discharge. Make a map and cross-sections of the data using flopy.plot methods. Plot specific discharge vectors on the map and cross-sections.
[57]:
head = gwf.output.head().get_data()
[58]:
gwf.output.budget().get_unique_record_names()
[58]:
[b' FLOW-JA-FACE',
b' DATA-SPDIS',
b' DATA-SAT',
b' WEL',
b' RCHA',
b' CHD',
b' SFR']
[59]:
spdis = gwf.output.budget().get_data(text="DATA-SPDIS")[0]
qx, qy, qz = flopy.utils.postprocessing.get_specific_discharge(
spdis, gwf, head=head
)
[60]:
fig, ax = plt.subplots(
ncols=1,
nrows=1,
figsize=(5, 8),
constrained_layout=True,
subplot_kw=dict(aspect="equal"),
)
mm = flopy.plot.PlotMapView(model=gwf, ax=ax, layer=0)
cb = mm.plot_array(head, vmin=10, vmax=30)
river.plot(color="cyan", ax=mm.ax)
mm.plot_grid(lw=0.5, color="0.5")
mm.plot_vector(qx, qy, qz, normalize=True)
cs = mm.contour_array(
head,
colors="red",
levels=np.arange(10, 28, 1),
linestyles=":",
linewidths=1.0,
tri_mask=True,
)
ax.clabel(
cs,
inline=True,
fmt="%1.0f",
fontsize=10,
inline_spacing=0.5,
)
plt.colorbar(cb, ax=mm.ax, shrink=0.5)
fig.savefig(output_folder / "freyberg-voronoi.png", dpi=300);
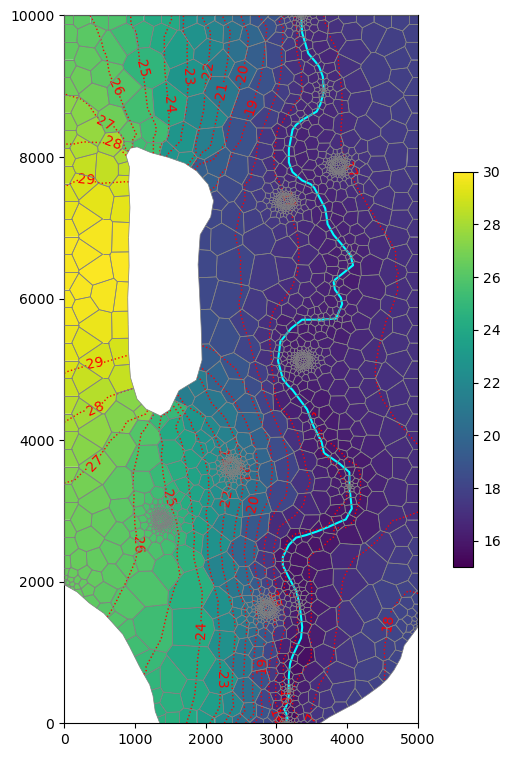
[61]:
fig, ax = plt.subplots(
ncols=1,
nrows=1,
figsize=(5, 8),
constrained_layout=True,
subplot_kw=dict(aspect="equal"),
)
mm = flopy.plot.PlotMapView(model=gwf, ax=ax, layer=0)
cb = mm.plot_array(gwf.dis.top.array, masked_values=[1e30], vmin=15, vmax=30)
river.plot(color="cyan", ax=mm.ax)
mm.plot_grid(lw=0.5, color="0.5", zorder=11)
cs = mm.contour_array(
gwf.dis.top.array,
colors="red",
levels=np.arange(15, 31, 1),
linestyles=":",
linewidths=1.0,
tri_mask=True,
)
ax.clabel(
cs,
inline=True,
fmt="%1.0f",
fontsize=10,
inline_spacing=0.5,
)
plt.colorbar(cb, ax=mm.ax, shrink=0.5)
fig.savefig(output_folder / "freyberg-voronoi-grid.png", dpi=300);
[ ]: