04: Modelgrid and intersection
The first part of this notebook is focused on the flopy.discretization.Grid (modelgrid) object(s). The modelgrid object(s) are a relatively new addition to FloPy’s capabilities and are the backbone of plotting, exporting, and GIS data processing within FloPy. These objects are automatically created when a model is loaded. Alternatively they can also be created as a stand alone object.
There are three types of modelgrids:
StructuredGrid: the StructuredGrid object is created for rectilinear grids. i.e. models that use a DIS file for discretizationVertexGrid: the VertexGrid object is for discretizations that are defined by vertices (e.g., DISV packages)UnstructuredGrid: the UnstructuredGrid object is for unstructured discretizations (e.g., DISU and MODFLOW-USG)
These objects all have a common interface defined by the base Grid class. What this means for the user is if they want to get the cell center coordinates from a modelgrid, the function call is identical on all three grids.
The best way to learn about these classes is by checking them out.
[1]:
from pathlib import Path
import flopy
from flopy.utils.gridgen import Gridgen
from flopy.utils import GridIntersect, Raster
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import geopandas as gpd
# pd.options.mode.chained_assignment = None
data_path = Path("../data/modelgrid_intersection")
Grids
StructuredGrid
Let’s start with the most common type of MODFLOW modelgrid, the StructuredGrid
First we’ll build one from scratch
[2]:
nrow = 20
ncol = 15
nlay = 1
grad = np.expand_dims(np.linspace(50, 100, nrow), axis=1)[::-1]
top = np.ones((nrow, ncol)) * grad
botm = np.zeros((nlay, nrow, ncol))
botm[0, :, :] = top - 50
delr = np.full((ncol,), 250)
delc = np.full((nrow,), 150)
ibound = np.ones((nlay, nrow, ncol))
ibound[0, 4:10, 4:8] = 0
modelgrid = flopy.discretization.StructuredGrid(
delc,
delr,
top,
botm,
ibound,
)
print(modelgrid)
print(type(modelgrid))
xll:0.0; yll:0.0; rotation:0.0; units:undefined; lenuni:0
<class 'flopy.discretization.structuredgrid.StructuredGrid'>
Let’s take a look at our modelgrid visually
[3]:
modelgrid.plot();
This is great, but what if we want to orient it in space and add a coordinate reference system so we can do further processing?
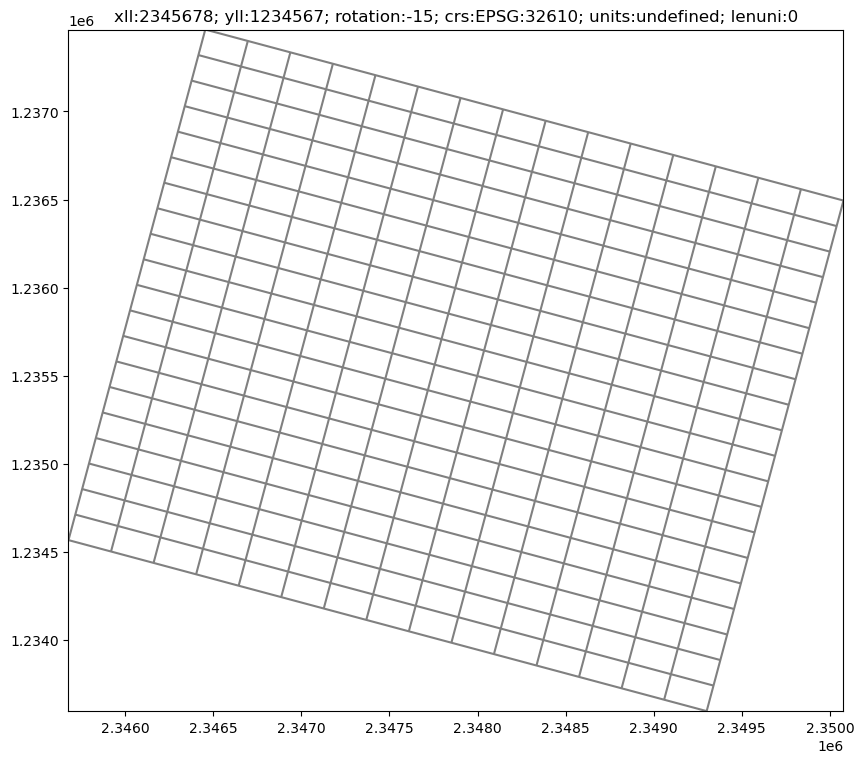
The set_coord_info() method allows us to update the coordinate offsets, rotation of the grid, and even add projection information.
[4]:
xoff = 2345678
yoff = 1234567
angrot = -15
epsg = 32610 # utm zone 10N
modelgrid.set_coord_info(xoff=xoff, yoff=yoff, angrot=angrot, crs=epsg)
[5]:
fig, ax = plt.subplots(figsize=(10, 10))
ax = modelgrid.plot(ax=ax)
plt.title(str(modelgrid));
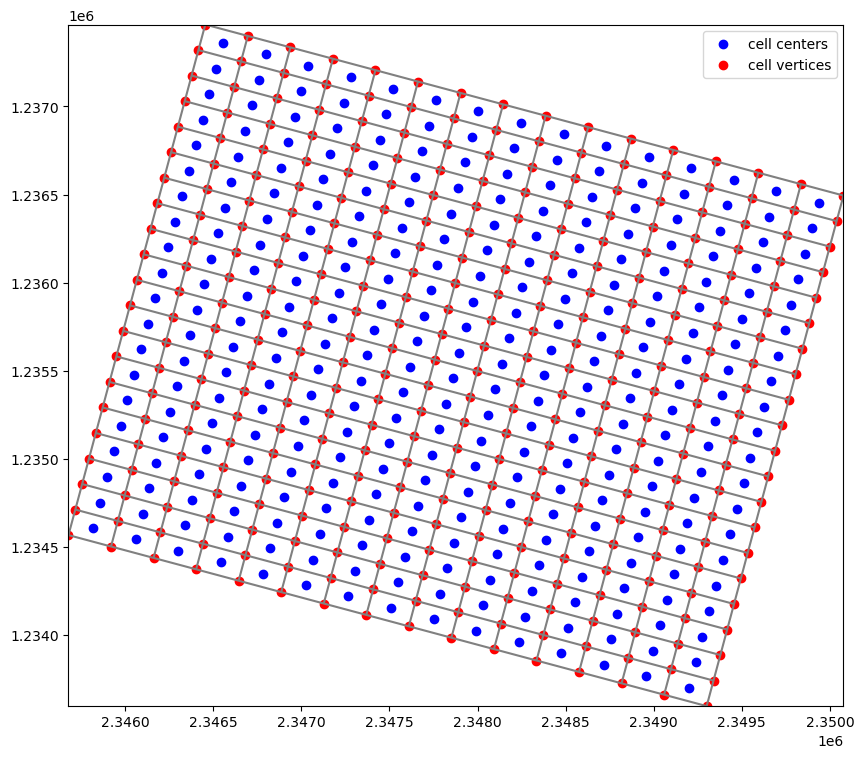
Getting cell centers and cell vertices
Cell centers can be returned to the user through the
.xcellcenters,.ycellcenter, and.zcellcenterproperties.Cell vertices can be returned to the user through the
.xverticesand.yverticesproperties
[6]:
xc, yc = modelgrid.xcellcenters, modelgrid.ycellcenters
xv, yv = modelgrid.xvertices, modelgrid.yvertices
fig, ax = plt.subplots(figsize=(10, 10))
modelgrid.plot()
ax.scatter(xc.ravel(), yc.ravel(), c="b", label="cell centers")
ax.scatter(xv.ravel(), yv.ravel(), c="r", label="cell vertices")
plt.legend(loc=0);
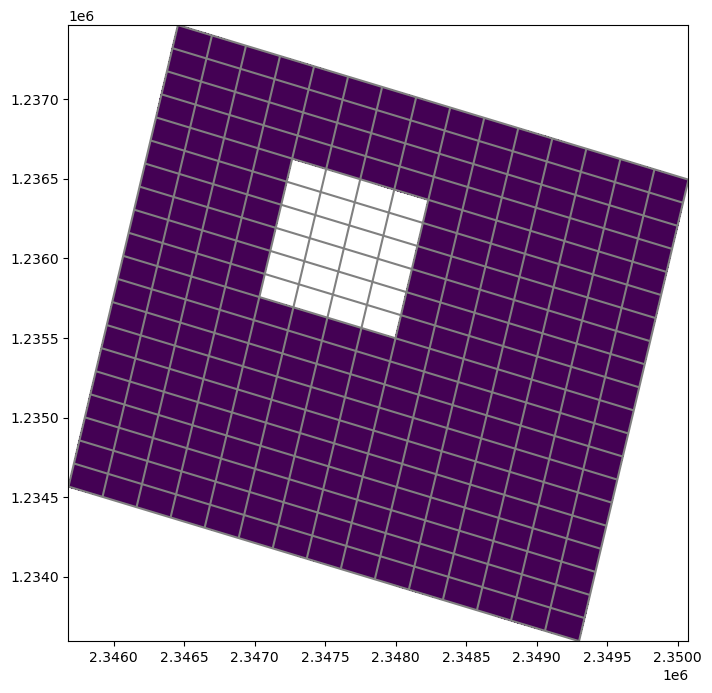
Getting the ibound/idomain array
The .idomain method returns the ibound/idomain array
[7]:
ibound = modelgrid.idomain
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(modelgrid=modelgrid, ax=ax)
pmv.plot_grid()
pmv.plot_array(ibound, masked_values=[0,]);
Getting the model top, bottom, delc, and delr
The model top, bottom, delc, and delr can be accessed from the modelgrid using .top, .bottom, .delc, and .delr.
[8]:
top = modelgrid.top
botm = modelgrid.botm
delc = modelgrid.delc
delr = modelgrid.delr
top, botm, delc, delr
[8]:
(array([[100. , 100. , 100. , 100. ,
100. , 100. , 100. , 100. ,
100. , 100. , 100. , 100. ,
100. , 100. , 100. ],
[ 97.36842105, 97.36842105, 97.36842105, 97.36842105,
97.36842105, 97.36842105, 97.36842105, 97.36842105,
97.36842105, 97.36842105, 97.36842105, 97.36842105,
97.36842105, 97.36842105, 97.36842105],
[ 94.73684211, 94.73684211, 94.73684211, 94.73684211,
94.73684211, 94.73684211, 94.73684211, 94.73684211,
94.73684211, 94.73684211, 94.73684211, 94.73684211,
94.73684211, 94.73684211, 94.73684211],
[ 92.10526316, 92.10526316, 92.10526316, 92.10526316,
92.10526316, 92.10526316, 92.10526316, 92.10526316,
92.10526316, 92.10526316, 92.10526316, 92.10526316,
92.10526316, 92.10526316, 92.10526316],
[ 89.47368421, 89.47368421, 89.47368421, 89.47368421,
89.47368421, 89.47368421, 89.47368421, 89.47368421,
89.47368421, 89.47368421, 89.47368421, 89.47368421,
89.47368421, 89.47368421, 89.47368421],
[ 86.84210526, 86.84210526, 86.84210526, 86.84210526,
86.84210526, 86.84210526, 86.84210526, 86.84210526,
86.84210526, 86.84210526, 86.84210526, 86.84210526,
86.84210526, 86.84210526, 86.84210526],
[ 84.21052632, 84.21052632, 84.21052632, 84.21052632,
84.21052632, 84.21052632, 84.21052632, 84.21052632,
84.21052632, 84.21052632, 84.21052632, 84.21052632,
84.21052632, 84.21052632, 84.21052632],
[ 81.57894737, 81.57894737, 81.57894737, 81.57894737,
81.57894737, 81.57894737, 81.57894737, 81.57894737,
81.57894737, 81.57894737, 81.57894737, 81.57894737,
81.57894737, 81.57894737, 81.57894737],
[ 78.94736842, 78.94736842, 78.94736842, 78.94736842,
78.94736842, 78.94736842, 78.94736842, 78.94736842,
78.94736842, 78.94736842, 78.94736842, 78.94736842,
78.94736842, 78.94736842, 78.94736842],
[ 76.31578947, 76.31578947, 76.31578947, 76.31578947,
76.31578947, 76.31578947, 76.31578947, 76.31578947,
76.31578947, 76.31578947, 76.31578947, 76.31578947,
76.31578947, 76.31578947, 76.31578947],
[ 73.68421053, 73.68421053, 73.68421053, 73.68421053,
73.68421053, 73.68421053, 73.68421053, 73.68421053,
73.68421053, 73.68421053, 73.68421053, 73.68421053,
73.68421053, 73.68421053, 73.68421053],
[ 71.05263158, 71.05263158, 71.05263158, 71.05263158,
71.05263158, 71.05263158, 71.05263158, 71.05263158,
71.05263158, 71.05263158, 71.05263158, 71.05263158,
71.05263158, 71.05263158, 71.05263158],
[ 68.42105263, 68.42105263, 68.42105263, 68.42105263,
68.42105263, 68.42105263, 68.42105263, 68.42105263,
68.42105263, 68.42105263, 68.42105263, 68.42105263,
68.42105263, 68.42105263, 68.42105263],
[ 65.78947368, 65.78947368, 65.78947368, 65.78947368,
65.78947368, 65.78947368, 65.78947368, 65.78947368,
65.78947368, 65.78947368, 65.78947368, 65.78947368,
65.78947368, 65.78947368, 65.78947368],
[ 63.15789474, 63.15789474, 63.15789474, 63.15789474,
63.15789474, 63.15789474, 63.15789474, 63.15789474,
63.15789474, 63.15789474, 63.15789474, 63.15789474,
63.15789474, 63.15789474, 63.15789474],
[ 60.52631579, 60.52631579, 60.52631579, 60.52631579,
60.52631579, 60.52631579, 60.52631579, 60.52631579,
60.52631579, 60.52631579, 60.52631579, 60.52631579,
60.52631579, 60.52631579, 60.52631579],
[ 57.89473684, 57.89473684, 57.89473684, 57.89473684,
57.89473684, 57.89473684, 57.89473684, 57.89473684,
57.89473684, 57.89473684, 57.89473684, 57.89473684,
57.89473684, 57.89473684, 57.89473684],
[ 55.26315789, 55.26315789, 55.26315789, 55.26315789,
55.26315789, 55.26315789, 55.26315789, 55.26315789,
55.26315789, 55.26315789, 55.26315789, 55.26315789,
55.26315789, 55.26315789, 55.26315789],
[ 52.63157895, 52.63157895, 52.63157895, 52.63157895,
52.63157895, 52.63157895, 52.63157895, 52.63157895,
52.63157895, 52.63157895, 52.63157895, 52.63157895,
52.63157895, 52.63157895, 52.63157895],
[ 50. , 50. , 50. , 50. ,
50. , 50. , 50. , 50. ,
50. , 50. , 50. , 50. ,
50. , 50. , 50. ]]),
array([[[50. , 50. , 50. , 50. ,
50. , 50. , 50. , 50. ,
50. , 50. , 50. , 50. ,
50. , 50. , 50. ],
[47.36842105, 47.36842105, 47.36842105, 47.36842105,
47.36842105, 47.36842105, 47.36842105, 47.36842105,
47.36842105, 47.36842105, 47.36842105, 47.36842105,
47.36842105, 47.36842105, 47.36842105],
[44.73684211, 44.73684211, 44.73684211, 44.73684211,
44.73684211, 44.73684211, 44.73684211, 44.73684211,
44.73684211, 44.73684211, 44.73684211, 44.73684211,
44.73684211, 44.73684211, 44.73684211],
[42.10526316, 42.10526316, 42.10526316, 42.10526316,
42.10526316, 42.10526316, 42.10526316, 42.10526316,
42.10526316, 42.10526316, 42.10526316, 42.10526316,
42.10526316, 42.10526316, 42.10526316],
[39.47368421, 39.47368421, 39.47368421, 39.47368421,
39.47368421, 39.47368421, 39.47368421, 39.47368421,
39.47368421, 39.47368421, 39.47368421, 39.47368421,
39.47368421, 39.47368421, 39.47368421],
[36.84210526, 36.84210526, 36.84210526, 36.84210526,
36.84210526, 36.84210526, 36.84210526, 36.84210526,
36.84210526, 36.84210526, 36.84210526, 36.84210526,
36.84210526, 36.84210526, 36.84210526],
[34.21052632, 34.21052632, 34.21052632, 34.21052632,
34.21052632, 34.21052632, 34.21052632, 34.21052632,
34.21052632, 34.21052632, 34.21052632, 34.21052632,
34.21052632, 34.21052632, 34.21052632],
[31.57894737, 31.57894737, 31.57894737, 31.57894737,
31.57894737, 31.57894737, 31.57894737, 31.57894737,
31.57894737, 31.57894737, 31.57894737, 31.57894737,
31.57894737, 31.57894737, 31.57894737],
[28.94736842, 28.94736842, 28.94736842, 28.94736842,
28.94736842, 28.94736842, 28.94736842, 28.94736842,
28.94736842, 28.94736842, 28.94736842, 28.94736842,
28.94736842, 28.94736842, 28.94736842],
[26.31578947, 26.31578947, 26.31578947, 26.31578947,
26.31578947, 26.31578947, 26.31578947, 26.31578947,
26.31578947, 26.31578947, 26.31578947, 26.31578947,
26.31578947, 26.31578947, 26.31578947],
[23.68421053, 23.68421053, 23.68421053, 23.68421053,
23.68421053, 23.68421053, 23.68421053, 23.68421053,
23.68421053, 23.68421053, 23.68421053, 23.68421053,
23.68421053, 23.68421053, 23.68421053],
[21.05263158, 21.05263158, 21.05263158, 21.05263158,
21.05263158, 21.05263158, 21.05263158, 21.05263158,
21.05263158, 21.05263158, 21.05263158, 21.05263158,
21.05263158, 21.05263158, 21.05263158],
[18.42105263, 18.42105263, 18.42105263, 18.42105263,
18.42105263, 18.42105263, 18.42105263, 18.42105263,
18.42105263, 18.42105263, 18.42105263, 18.42105263,
18.42105263, 18.42105263, 18.42105263],
[15.78947368, 15.78947368, 15.78947368, 15.78947368,
15.78947368, 15.78947368, 15.78947368, 15.78947368,
15.78947368, 15.78947368, 15.78947368, 15.78947368,
15.78947368, 15.78947368, 15.78947368],
[13.15789474, 13.15789474, 13.15789474, 13.15789474,
13.15789474, 13.15789474, 13.15789474, 13.15789474,
13.15789474, 13.15789474, 13.15789474, 13.15789474,
13.15789474, 13.15789474, 13.15789474],
[10.52631579, 10.52631579, 10.52631579, 10.52631579,
10.52631579, 10.52631579, 10.52631579, 10.52631579,
10.52631579, 10.52631579, 10.52631579, 10.52631579,
10.52631579, 10.52631579, 10.52631579],
[ 7.89473684, 7.89473684, 7.89473684, 7.89473684,
7.89473684, 7.89473684, 7.89473684, 7.89473684,
7.89473684, 7.89473684, 7.89473684, 7.89473684,
7.89473684, 7.89473684, 7.89473684],
[ 5.26315789, 5.26315789, 5.26315789, 5.26315789,
5.26315789, 5.26315789, 5.26315789, 5.26315789,
5.26315789, 5.26315789, 5.26315789, 5.26315789,
5.26315789, 5.26315789, 5.26315789],
[ 2.63157895, 2.63157895, 2.63157895, 2.63157895,
2.63157895, 2.63157895, 2.63157895, 2.63157895,
2.63157895, 2.63157895, 2.63157895, 2.63157895,
2.63157895, 2.63157895, 2.63157895],
[ 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. ,
0. , 0. , 0. ]]]),
array([150., 150., 150., 150., 150., 150., 150., 150., 150., 150., 150.,
150., 150., 150., 150., 150., 150., 150., 150., 150.]),
array([250., 250., 250., 250., 250., 250., 250., 250., 250., 250., 250.,
250., 250., 250., 250.]))
Class exercise 1:
Assume that the modelgrid is currently in meters and we want our grid to be in feet. Create a new modelgrid object from the existing one where the discretization is in feet.
make a plot of the new modelgrid after it has been created
[9]:
conv = 3.28084
mg2 = flopy.discretization.StructuredGrid(
modelgrid.delc * conv,
modelgrid.delr * conv,
modelgrid.top * conv,
modelgrid.botm * conv,
modelgrid.idomain,
xoff=xoff * conv,
yoff=yoff * conv,
angrot=angrot
)
print(mg2)
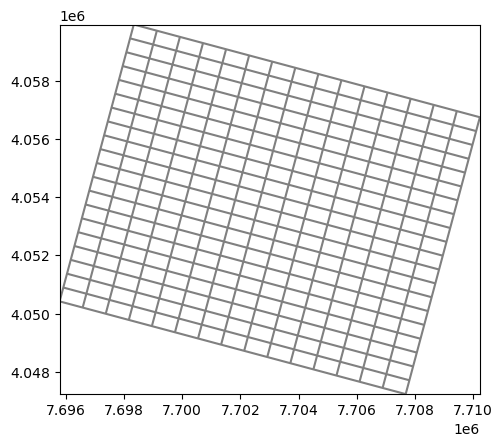
mg2.plot();
xll:7695794.20952; yll:4050416.79628; rotation:-15; units:undefined; lenuni:0
Imagine that we want to find the nearest neighbors of a cell for some reason. The modelgrid has a built in method to 1) internally construct a table of nearest neighbors and 2) return the nearest neighbors for a given cell to the user.
[10]:
cell = (0, 0, 5)
neighbors = modelgrid.neighbors(*cell)
neighbors
[10]:
[(np.int64(0), np.int64(0), np.int64(4)),
(np.int64(0), np.int64(0), np.int64(6)),
(np.int64(0), np.int64(1), np.int64(5))]
If we want to find a cell number based on x, y coordinate location the model grid has a built in .intersect() method that takes x, y, and an optional z coordinate and returns the cell number
[11]:
x, y, z = 2348000, 1235000, 25
rowcol = modelgrid.intersect(x, y) # without z
layrowcol = modelgrid.intersect(x, y, z) # with z location
f"{rowcol=}, {layrowcol=}"
[11]:
'rowcol=(np.int64(13), np.int64(8)), layrowcol=(0, np.int64(13), np.int64(8))'
Vertex model grid
The vertex model grid is produced for MODFLOW model discretizations that are defined by vertices (DISV). We can build a quadtree vertex grid from an exiting structured modelgrid using the gridgen utility and the Gridgen class.
First let’s unrotate the grid coordinates of our structured grid to make this easy
[12]:
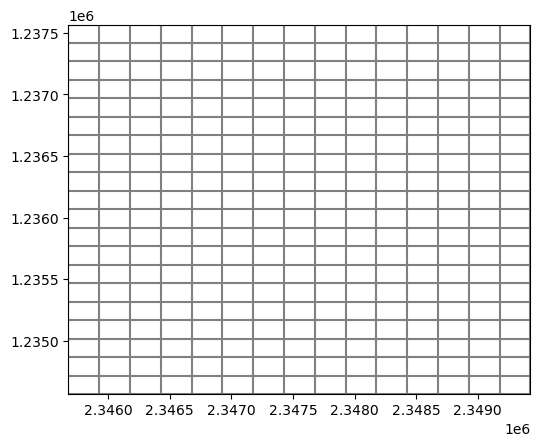
modelgrid.set_coord_info(angrot=0)
modelgrid.plot();
Lets load a couple of shapefiles to help with creating the quadtree
[13]:
active_shp = data_path / "active_area.shp"
refine_shp = data_path / "refined_area.shp"
active = gpd.read_file(active_shp)
refined = gpd.read_file(refine_shp)
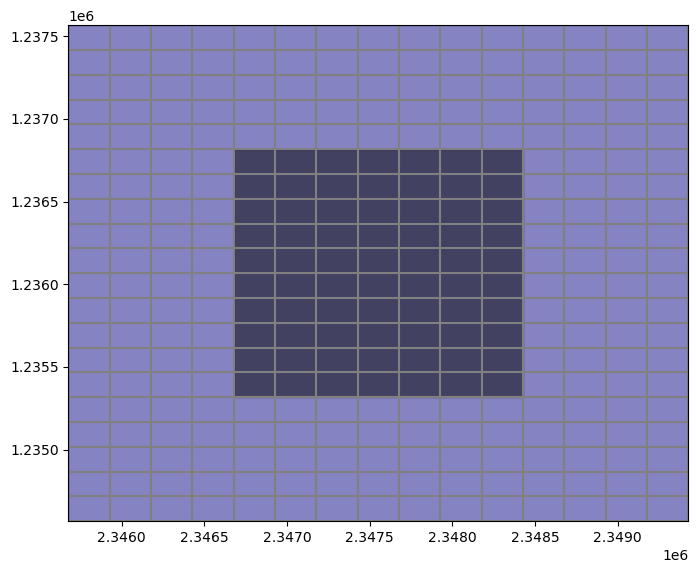
Now let’s plot these areas on the grid to visualize them
[14]:
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(modelgrid=modelgrid)
pmv.plot_grid()
# active.geometry.to_list()
pmv.plot_shapes(active.geometry.to_list(), alpha=0.5, cmap='plasma')
pmv.plot_shapes(refined.geometry.to_list(), alpha=0.5, cmap='magma');
[15]:
g = Gridgen(modelgrid, model_ws=data_path)
g.add_refinement_features(refined.geometry.to_list(), "polygon", 1, [0,])
g.build(verbose=False)
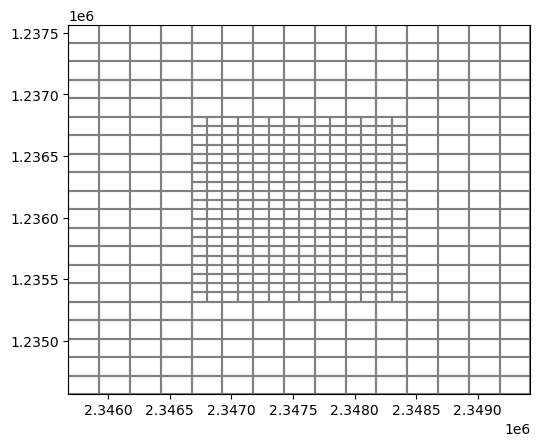
Create a vertex model grid from the results
[16]:
gridprops = g.get_gridprops_vertexgrid()
vertexgrid = flopy.discretization.VertexGrid(**gridprops)
vertexgrid.plot();
Getting the modelgrid’s shape
[17]:
shape = vertexgrid.shape
shape
[17]:
(1, 510)
Class Exercise 2:
Find and plot all of the nearest neighbors at node 79 on the vertex modelgrid.
Hint: Use flopy.plot.PlotMapView for your plotting, and plot a boolean (1, 0) array with the cells that are not neighbors masked out.
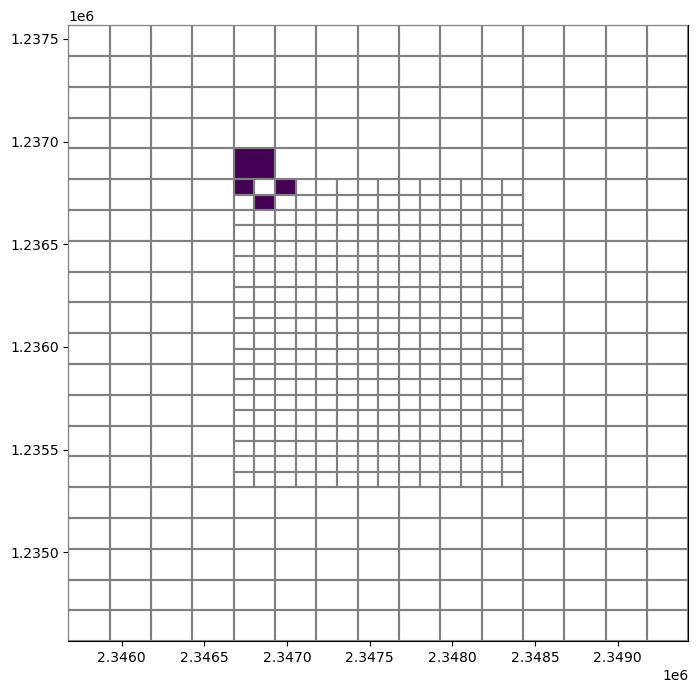
[18]:
neighbors = vertexgrid.neighbors(80)
arr = np.zeros((vertexgrid.shape[-1]), dtype=int)
arr[neighbors] = 1
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(ax=ax, modelgrid=vertexgrid)
pmv.plot_grid(zorder=3)
pmv.plot_array(arr, masked_values=[0,]);
UnstructuredGrid
The UnstructuredGrid class is used with MODFLOW-USG and MODFLOW-6 DISU type discretizations. We can also create an UnstructuredGrid object from the Gridgen results
[19]:
gridprops = g.get_gridprops_unstructuredgrid()
ugrid = flopy.discretization.UnstructuredGrid(**gridprops)
ugrid, type(ugrid)
[19]:
(xll:0.0; yll:0.0; rotation:0.0; units:undefined; lenuni:0,
flopy.discretization.unstructuredgrid.UnstructuredGrid)
[20]:
ugrid.plot();
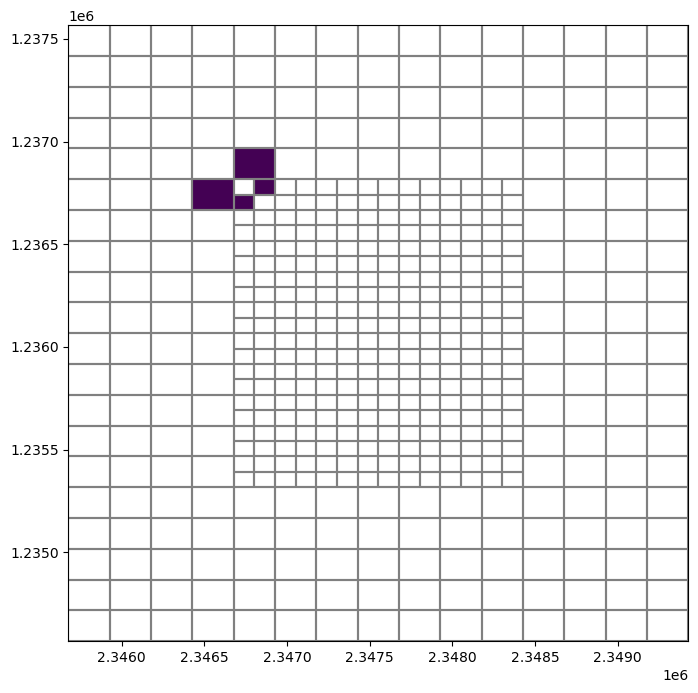
[21]:
neighbors = ugrid.neighbors(79)
arr = np.zeros(ugrid.shape, dtype=int)
arr[neighbors] = 1
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(ax=ax, modelgrid=ugrid)
pmv.plot_grid()
pmv.plot_array(arr, masked_values=[0,]);
Intersecting geospatial data
In this part of the lesson we will intersect geospatial data with a modelgrid and begin building a simple adaptation of the Sagehen Creek model, near Truckee, CA. While building this model, we’ll work with shapefile and raster data. As we build the model FloPy’s intersection capabilities will be presented.
Let’s start by getting the basin boundary. For the Sagehen Creek model, we’re interested in the contributing area for USGS gage 10343500
[22]:
station_id = "10343500"
basin = gpd.read_file(data_path / "sagehen_basin.shp")
basin = basin.set_index("identifier")
basin
[22]:
| geometry | |
|---|---|
| identifier | |
| 10343500 | POLYGON ((-120.25104 39.40471, -120.25613 39.4... |
First thing that we notice is that the basin polygon is projected in decimal degrees, which is not good for modeling. An equal area projection would be better suited for this application
[23]:
epsg = 26911 # NAD83 utm zone 11 N, epsg: 26911
basin = basin.to_crs(epsg=epsg)
basin
[23]:
| geometry | |
|---|---|
| identifier | |
| 10343500 | POLYGON ((220068.76 4366733.474, 219647.52 436... |
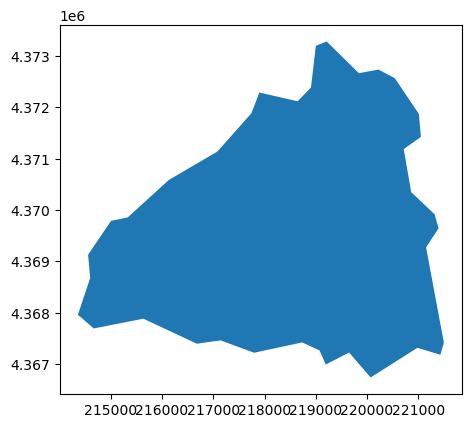
[24]:
basin.plot()
[24]:
<Axes: >
We can now get our basin’s bounding box using .bounds, buffer it, and then begin creating a modelgrid based on the basin boundary.
[25]:
cellsize = 90 # 90 m grid
bounds = basin.bounds
print(bounds)
xmin = bounds.loc[station_id, "minx"] - (cellsize * 1)
ymin = bounds.loc[station_id, "miny"] - (cellsize * 1)
xmax = bounds.loc[station_id, "maxx"] + (cellsize * 1)
ymax = bounds.loc[station_id, "maxy"] + (cellsize * 1)
minx miny maxx maxy
identifier
10343500 214358.622076 4.366733e+06 221497.557955 4.373287e+06
Calculate the number of rows and columns the model will have and then create a delc and delr array.
[26]:
dx = xmax - xmin
dy = ymax - ymin
nlay = 1
nrow = np.ceil(dy / cellsize).astype(int)
ncol = np.ceil(dx / cellsize).astype(int)
delr = np.full((ncol,), cellsize)
delc = np.full((nrow,), cellsize)
# let's create a fake top and bottom for now
top = np.full((nrow, ncol), 100)
botm = np.full((nlay, nrow, ncol), 0)
Now we can create a minimal modelgrid from the information we have
[27]:
modelgrid = flopy.discretization.StructuredGrid(
delc,
delr,
top=top,
botm=botm,
nlay=nlay,
xoff=xmin,
yoff=ymin,
crs=epsg
)
print(modelgrid)
print(modelgrid.is_complete)
xll:214268.62207605218; yll:4366643.474127463; rotation:0.0; crs:EPSG:26911; units:undefined; lenuni:0
False
[28]:
fig, ax = plt.subplots(figsize=(8, 8))
ax = basin.plot(ax=ax)
modelgrid.plot(ax=ax, lw=0.5);
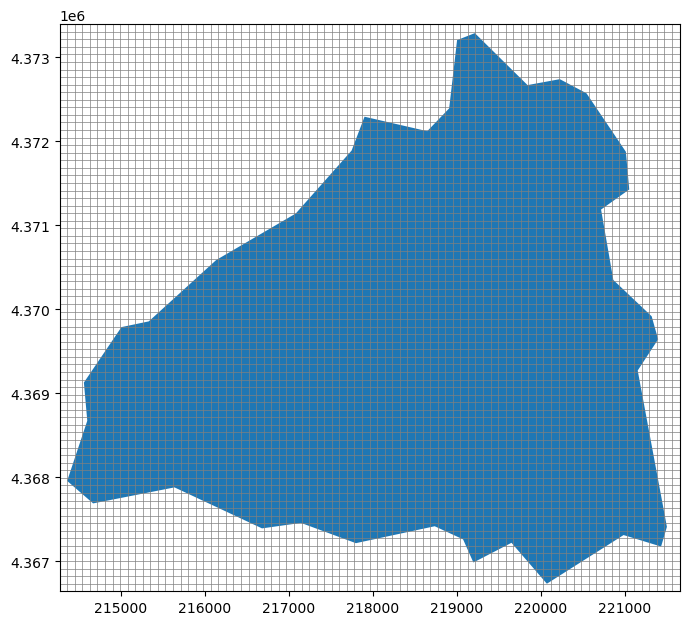
Fantastic! The modelgrid has been created and it completely overlays the contributing area.
FloPy has a utility named GridIntersect that allows the user to intersect points, polygons, and polylines with modelgrid objects.
In this example, the basin boundary will be intersected with the modelgrid to create the active and inactive extents of the model. The .intersect() method returns a numpy recarry with cellids, vertices, cell areas, and shapely polygon objects.
[29]:
gx = GridIntersect(modelgrid)
result = gx.intersect(basin.loc[station_id, "geometry"])
result.cellids
/Users/mnfienen/miniforge3/envs/pyclass/lib/python3.11/site-packages/flopy/utils/gridintersect.py:123: DeprecationWarning: Note `method="structured"` is deprecated. Pass `method="vertex"` to silence this warning. This will be the new default in a future release and this keyword argument will be removed.
warnings.warn(
[29]:
array([(1, 52), (1, 53), (1, 54), ..., (73, 64), (73, 65), (73, 66)],
shape=(3587,), dtype=object)
create the ibound/idomain array (active and inactive model cells)
[30]:
ibound = np.zeros((nlay, nrow, ncol))
i, j = zip(*result.cellids)
ibound[:, i, j] = 1
fig, ax = plt.subplots(figsize=(8, 8))
pmv = flopy.plot.PlotMapView(modelgrid=modelgrid, ax=ax)
pmv.plot_grid(lw=0.5)
pmv.plot_array(ibound, masked_values=[0], cmap="viridis")
[30]:
<matplotlib.collections.QuadMesh at 0x165463290>
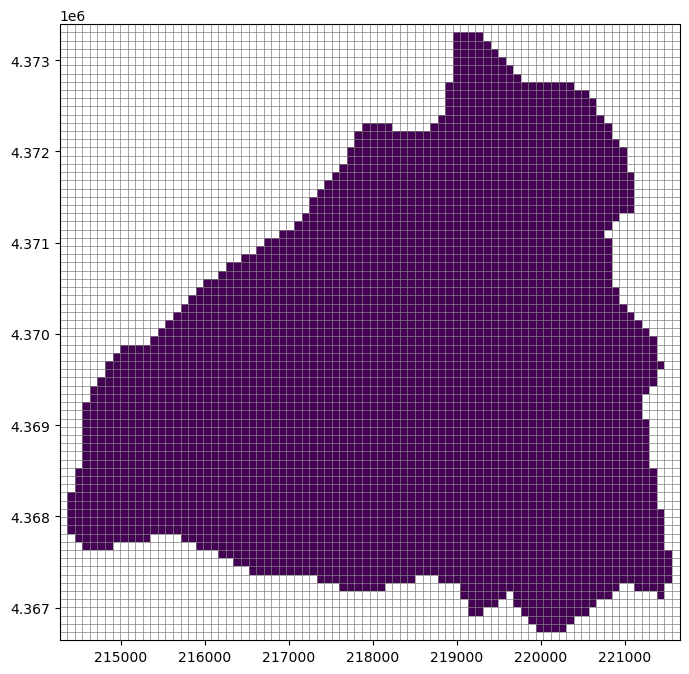
FloPy’s Raster class allows us to intersect and resample raster data to the model grid. This class is useful for resampling a number of gridded products such as: Digital Elevation Model data, land cover data, and gridded climate data.
The first example of using the Raster class will be to resample Digital Elevation Model (DEM) data to define the land surface of the Sagehen model
[31]:
dem_file = data_path / "dem_30m.img"
raster = Raster.load(dem_file)
Now that the raster is loaded we can resample it
The resample_to_grid() method performs geostatistics and returns an array in the shape of the modelgrid. Options for resampling include "nearest", "linear" (bilinear), "cubic" (bicubic), "max", "min", "mean", "median", and "mode" (most common value).
[32]:
# let's get the nearest neighbor elevations
dem_data = raster.resample_to_grid(
modelgrid,
band=raster.bands[0],
method="nearest"
)
dem_data
[32]:
array([[2288.88427734, 2273.79296875, 2263.59692383, ..., 2035.27868652,
2020.77258301, 2011.82592773],
[2308.66918945, 2301.82080078, 2284.20483398, ..., 2037.7668457 ,
2022.42944336, 2011.48937988],
[2326.71118164, 2321.3112793 , 2301.70703125, ..., 2042.32263184,
2025.94592285, 2015.38793945],
...,
[2207.31152344, 2229.87426758, 2239.453125 , ..., 2112.20751953,
2095.49414062, 2073.16308594],
[2176.74121094, 2186.13769531, 2201.04858398, ..., 2095.2331543 ,
2074.06396484, 2051.18530273],
[2156.98022461, 2157.87744141, 2164.14770508, ..., 2077.35131836,
2053.95043945, 2035.44116211]], shape=(75, 82))
[33]:
# Now let's plot it up and see what we have
fig, ax = plt.subplots(figsize=(10, 8))
pmv = flopy.plot.PlotMapView(ax=ax, modelgrid=modelgrid)
pmv.plot_grid(lw=0.5, zorder=3)
pc = pmv.plot_array(dem_data, alpha=0.75, zorder=1, cmap="viridis")
pmv.plot_array(ibound, masked_values=[1], cmap="magma", zorder=2)
plt.colorbar(pc, shrink=0.7);
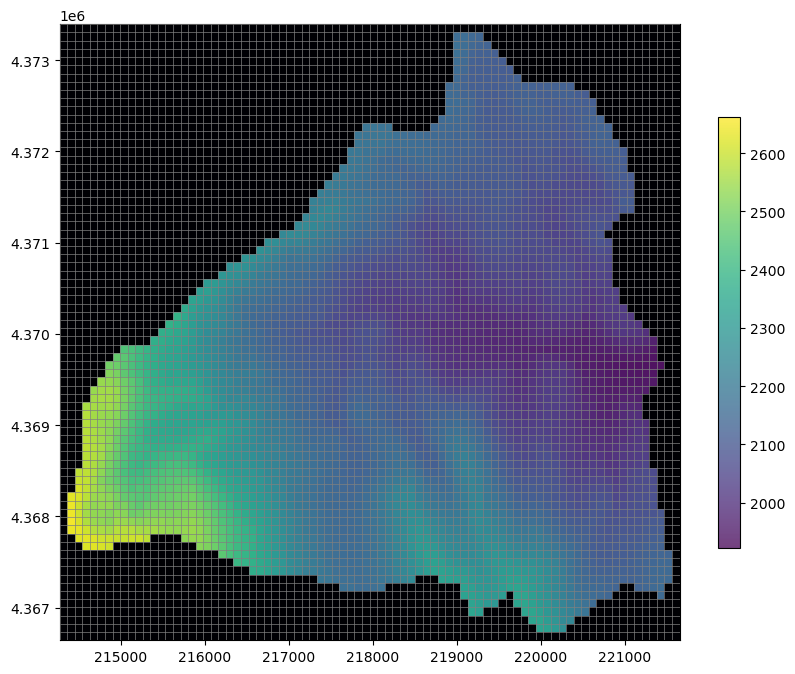
Start creating model files from the data we’ve gathered so far.
At this point we have almost everything we need to begin creating a model discretization package. The only thing left to calculate is the model bottom. Let’s assume bedrock 150 m below land surface throughout the basin and begin creating our model.
We’ll first start by creating our simulation object and adding a solver and the time discretization. This model will be a year long with 12 stress periods and a daily time step
[34]:
sim = flopy.mf6.MFSimulation("sagehen", sim_ws=data_path)
ims = flopy.mf6.ModflowIms(sim, complexity="COMPLEX")
nper = 12
perlen = (31, 28, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31)
period_data = [(i, i, 1.0) for i in perlen]
tdis = flopy.mf6.ModflowTdis(
sim,
nper=12,
perioddata=period_data,
time_units="days"
)
Now to create the Groundwater flow model object and add the dis file to it
[35]:
gwf = flopy.mf6.ModflowGwf(
sim,
modelname='sagehen',
save_flows=True,
)
# add the dis package
botm = np.expand_dims(dem_data, axis=0) - 150
dis = flopy.mf6.ModflowGwfdis(
gwf,
nrow=modelgrid.nrow,
ncol=modelgrid.ncol,
delr=modelgrid.delr,
delc=modelgrid.delc,
top=dem_data,
botm=botm,
idomain=ibound
)
gwf
[35]:
name = sagehen
model_type = gwf6
version = mf6
model_relative_path = .
###################
Package dis
###################
package_name = dis
filename = sagehen.dis
package_type = dis
model_or_simulation_package = model
model_name = sagehen
We can also add an initial conditions file to the model. Let’s assume water level begins at 5 meters below land surface
[36]:
ic = flopy.mf6.ModflowGwfic(gwf, strt=dem_data - 5)
gwf
[36]:
name = sagehen
model_type = gwf6
version = mf6
model_relative_path = .
###################
Package dis
###################
package_name = dis
filename = sagehen.dis
package_type = dis
model_or_simulation_package = model
model_name = sagehen
###################
Package ic
###################
package_name = ic
filename = sagehen.ic
package_type = ic
model_or_simulation_package = model
model_name = sagehen
And we can create our NPF and STO packages using values from literature
[37]:
npf = flopy.mf6.ModflowGwfnpf(
gwf,
save_flows=True,
save_specific_discharge=True,
k=0.022 # value from Larsen et al, 2022
)
sto = flopy.mf6.ModflowGwfsto(
gwf,
save_flows=True,
iconvert=1,
ss=1e-07, # values from Larsen et al, 2022
sy=0.2
)
We can also add a OC package now
[38]:
# build output control package
budget_file = "sagehen.cbc"
head_file = "sagehen.hds"
saverecord = {i: [("HEAD", "ALL"), ("BUDGET", "ALL")] for i in range(gwf.nper)}
oc = flopy.mf6.ModflowGwfoc(
gwf,
budget_filerecord=budget_file,
head_filerecord=head_file,
saverecord=saverecord,
)
Class Exercise: Loading and resampling rasters
For this exercise we are loading and resampling monthly Daymet climate data for Sagehen basin.
Load the potential evapotranspiration raster pet_raster using flopy’s Raster class and create a dictionary of resampled PET that stores all 12 months of values.
Hints:
Rasters contain multiple datasets called bands, the
Rasterclass has a.bandsattribute that returns a list of all of the bands within the raster. Lists can be looped over!Because the data is coarse compared to our model discretization, the nearest neighbor method should be appropriate for resampling
method="nearest"Finally no data values should be converted to 0 value. The
Rasterclass has a.nodatavalsattribute that returns a list of no data values. There is only one in this list; use numpy’s boolean indexing ornp.where()to replace all of the no data entries with 0.
[39]:
pet_raster = data_path / "pet.tif"
prcp_raster = data_path / "prcp.tif"
[40]:
rstr = flopy.utils.Raster.load(pet_raster)
[41]:
pet_monthly = {}
for band in rstr.bands:
t = rstr.resample_to_grid(
modelgrid,
band,
method="nearest"
)
t[t == rstr.nodatavals[0]] = 0
pet_monthly[band - 1] = t
pet_monthly
[41]:
{0: array([[0.89827865, 0.89827865, 0.89827865, ..., 1.00581992, 1.00581992,
1.00581992],
[0.89827865, 0.89827865, 0.89827865, ..., 1.00581992, 1.00581992,
1.00581992],
[0.89827865, 0.89827865, 0.89827865, ..., 1.00581992, 1.00581992,
1.00581992],
...,
[0. , 0. , 0. , ..., 1.01081669, 1.01081669,
1.01081669],
[0. , 0. , 0. , ..., 1.01081669, 1.01081669,
1.01081669],
[0. , 0. , 0. , ..., 1.01081669, 1.01081669,
1.01081669]], shape=(75, 82)),
1: array([[1.31099296, 1.31099296, 1.31099296, ..., 1.42294502, 1.42294502,
1.42294502],
[1.31099296, 1.31099296, 1.31099296, ..., 1.42294502, 1.42294502,
1.42294502],
[1.31099296, 1.31099296, 1.31099296, ..., 1.42294502, 1.42294502,
1.42294502],
...,
[0. , 0. , 0. , ..., 1.4494915 , 1.4494915 ,
1.4494915 ],
[0. , 0. , 0. , ..., 1.4494915 , 1.4494915 ,
1.4494915 ],
[0. , 0. , 0. , ..., 1.4494915 , 1.4494915 ,
1.4494915 ]], shape=(75, 82)),
2: array([[1.88199592, 1.88199592, 1.88199592, ..., 2.01319814, 2.01319814,
2.01319814],
[1.88199592, 1.88199592, 1.88199592, ..., 2.01319814, 2.01319814,
2.01319814],
[1.88199592, 1.88199592, 1.88199592, ..., 2.01319814, 2.01319814,
2.01319814],
...,
[0. , 0. , 0. , ..., 2.06212258, 2.06212258,
2.06212258],
[0. , 0. , 0. , ..., 2.06212258, 2.06212258,
2.06212258],
[0. , 0. , 0. , ..., 2.06212258, 2.06212258,
2.06212258]], shape=(75, 82)),
3: array([[3.14511275, 3.14511275, 3.14511275, ..., 3.33968782, 3.33968782,
3.33968782],
[3.14511275, 3.14511275, 3.14511275, ..., 3.33968782, 3.33968782,
3.33968782],
[3.14511275, 3.14511275, 3.14511275, ..., 3.33968782, 3.33968782,
3.33968782],
...,
[0. , 0. , 0. , ..., 3.41348195, 3.41348195,
3.41348195],
[0. , 0. , 0. , ..., 3.41348195, 3.41348195,
3.41348195],
[0. , 0. , 0. , ..., 3.41348195, 3.41348195,
3.41348195]], shape=(75, 82)),
4: array([[3.94825053, 3.94825053, 3.94825053, ..., 4.04690742, 4.04690742,
4.04690742],
[3.94825053, 3.94825053, 3.94825053, ..., 4.04690742, 4.04690742,
4.04690742],
[3.94825053, 3.94825053, 3.94825053, ..., 4.04690742, 4.04690742,
4.04690742],
...,
[0. , 0. , 0. , ..., 4.1060009 , 4.1060009 ,
4.1060009 ],
[0. , 0. , 0. , ..., 4.1060009 , 4.1060009 ,
4.1060009 ],
[0. , 0. , 0. , ..., 4.1060009 , 4.1060009 ,
4.1060009 ]], shape=(75, 82)),
5: array([[4.99552679, 4.99552679, 4.99552679, ..., 5.388381 , 5.388381 ,
5.388381 ],
[4.99552679, 4.99552679, 4.99552679, ..., 5.388381 , 5.388381 ,
5.388381 ],
[4.99552679, 4.99552679, 4.99552679, ..., 5.388381 , 5.388381 ,
5.388381 ],
...,
[0. , 0. , 0. , ..., 5.34579992, 5.34579992,
5.34579992],
[0. , 0. , 0. , ..., 5.34579992, 5.34579992,
5.34579992],
[0. , 0. , 0. , ..., 5.34579992, 5.34579992,
5.34579992]], shape=(75, 82)),
6: array([[5.21027994, 5.21027994, 5.21027994, ..., 5.60202217, 5.60202217,
5.60202217],
[5.21027994, 5.21027994, 5.21027994, ..., 5.60202217, 5.60202217,
5.60202217],
[5.21027994, 5.21027994, 5.21027994, ..., 5.60202217, 5.60202217,
5.60202217],
...,
[0. , 0. , 0. , ..., 5.58002138, 5.58002138,
5.58002138],
[0. , 0. , 0. , ..., 5.58002138, 5.58002138,
5.58002138],
[0. , 0. , 0. , ..., 5.58002138, 5.58002138,
5.58002138]], shape=(75, 82)),
7: array([[4.54519796, 4.54519796, 4.54519796, ..., 4.94014788, 4.94014788,
4.94014788],
[4.54519796, 4.54519796, 4.54519796, ..., 4.94014788, 4.94014788,
4.94014788],
[4.54519796, 4.54519796, 4.54519796, ..., 4.94014788, 4.94014788,
4.94014788],
...,
[0. , 0. , 0. , ..., 4.97207499, 4.97207499,
4.97207499],
[0. , 0. , 0. , ..., 4.97207499, 4.97207499,
4.97207499],
[0. , 0. , 0. , ..., 4.97207499, 4.97207499,
4.97207499]], shape=(75, 82)),
8: array([[3.38186955, 3.38186955, 3.38186955, ..., 3.73335552, 3.73335552,
3.73335552],
[3.38186955, 3.38186955, 3.38186955, ..., 3.73335552, 3.73335552,
3.73335552],
[3.38186955, 3.38186955, 3.38186955, ..., 3.73335552, 3.73335552,
3.73335552],
...,
[0. , 0. , 0. , ..., 3.75254512, 3.75254512,
3.75254512],
[0. , 0. , 0. , ..., 3.75254512, 3.75254512,
3.75254512],
[0. , 0. , 0. , ..., 3.75254512, 3.75254512,
3.75254512]], shape=(75, 82)),
9: array([[1.60437179, 1.60437179, 1.60437179, ..., 1.80872381, 1.80872381,
1.80872381],
[1.60437179, 1.60437179, 1.60437179, ..., 1.80872381, 1.80872381,
1.80872381],
[1.60437179, 1.60437179, 1.60437179, ..., 1.80872381, 1.80872381,
1.80872381],
...,
[0. , 0. , 0. , ..., 1.84118676, 1.84118676,
1.84118676],
[0. , 0. , 0. , ..., 1.84118676, 1.84118676,
1.84118676],
[0. , 0. , 0. , ..., 1.84118676, 1.84118676,
1.84118676]], shape=(75, 82)),
10: array([[1.23011148, 1.23011148, 1.23011148, ..., 1.41456532, 1.41456532,
1.41456532],
[1.23011148, 1.23011148, 1.23011148, ..., 1.41456532, 1.41456532,
1.41456532],
[1.23011148, 1.23011148, 1.23011148, ..., 1.41456532, 1.41456532,
1.41456532],
...,
[0. , 0. , 0. , ..., 1.42739856, 1.42739856,
1.42739856],
[0. , 0. , 0. , ..., 1.42739856, 1.42739856,
1.42739856],
[0. , 0. , 0. , ..., 1.42739856, 1.42739856,
1.42739856]], shape=(75, 82)),
11: array([[0.76984066, 0.76984066, 0.76984066, ..., 0.85954601, 0.85954601,
0.85954601],
[0.76984066, 0.76984066, 0.76984066, ..., 0.85954601, 0.85954601,
0.85954601],
[0.76984066, 0.76984066, 0.76984066, ..., 0.85954601, 0.85954601,
0.85954601],
...,
[0. , 0. , 0. , ..., 0.8547039 , 0.8547039 ,
0.8547039 ],
[0. , 0. , 0. , ..., 0.8547039 , 0.8547039 ,
0.8547039 ],
[0. , 0. , 0. , ..., 0.8547039 , 0.8547039 ,
0.8547039 ]], shape=(75, 82))}
Let’s load the saved version of this data, it should be identical to the data that was produced in the exercise.
[42]:
arr = np.genfromtxt(data_path / "pet.txt")
arr.shape = (12, modelgrid.nrow, modelgrid.ncol)
pet_monthly = {i: a for i, a in enumerate(arr)}
pet_monthly
[42]:
{0: array([[0.89827865, 0.89827865, 0.89827865, ..., 1.00581992, 1.00581992,
1.00581992],
[0.89827865, 0.89827865, 0.89827865, ..., 1.00581992, 1.00581992,
1.00581992],
[0.89827865, 0.89827865, 0.89827865, ..., 1.00581992, 1.00581992,
1.00581992],
...,
[0. , 0. , 0. , ..., 1.01081669, 1.01081669,
1.01081669],
[0. , 0. , 0. , ..., 1.01081669, 1.01081669,
1.01081669],
[0. , 0. , 0. , ..., 1.01081669, 1.01081669,
1.01081669]], shape=(75, 82)),
1: array([[1.31099296, 1.31099296, 1.31099296, ..., 1.42294502, 1.42294502,
1.42294502],
[1.31099296, 1.31099296, 1.31099296, ..., 1.42294502, 1.42294502,
1.42294502],
[1.31099296, 1.31099296, 1.31099296, ..., 1.42294502, 1.42294502,
1.42294502],
...,
[0. , 0. , 0. , ..., 1.4494915 , 1.4494915 ,
1.4494915 ],
[0. , 0. , 0. , ..., 1.4494915 , 1.4494915 ,
1.4494915 ],
[0. , 0. , 0. , ..., 1.4494915 , 1.4494915 ,
1.4494915 ]], shape=(75, 82)),
2: array([[1.88199592, 1.88199592, 1.88199592, ..., 2.01319814, 2.01319814,
2.01319814],
[1.88199592, 1.88199592, 1.88199592, ..., 2.01319814, 2.01319814,
2.01319814],
[1.88199592, 1.88199592, 1.88199592, ..., 2.01319814, 2.01319814,
2.01319814],
...,
[0. , 0. , 0. , ..., 2.06212258, 2.06212258,
2.06212258],
[0. , 0. , 0. , ..., 2.06212258, 2.06212258,
2.06212258],
[0. , 0. , 0. , ..., 2.06212258, 2.06212258,
2.06212258]], shape=(75, 82)),
3: array([[3.14511275, 3.14511275, 3.14511275, ..., 3.33968782, 3.33968782,
3.33968782],
[3.14511275, 3.14511275, 3.14511275, ..., 3.33968782, 3.33968782,
3.33968782],
[3.14511275, 3.14511275, 3.14511275, ..., 3.33968782, 3.33968782,
3.33968782],
...,
[0. , 0. , 0. , ..., 3.41348195, 3.41348195,
3.41348195],
[0. , 0. , 0. , ..., 3.41348195, 3.41348195,
3.41348195],
[0. , 0. , 0. , ..., 3.41348195, 3.41348195,
3.41348195]], shape=(75, 82)),
4: array([[3.94825053, 3.94825053, 3.94825053, ..., 4.04690742, 4.04690742,
4.04690742],
[3.94825053, 3.94825053, 3.94825053, ..., 4.04690742, 4.04690742,
4.04690742],
[3.94825053, 3.94825053, 3.94825053, ..., 4.04690742, 4.04690742,
4.04690742],
...,
[0. , 0. , 0. , ..., 4.1060009 , 4.1060009 ,
4.1060009 ],
[0. , 0. , 0. , ..., 4.1060009 , 4.1060009 ,
4.1060009 ],
[0. , 0. , 0. , ..., 4.1060009 , 4.1060009 ,
4.1060009 ]], shape=(75, 82)),
5: array([[4.99552679, 4.99552679, 4.99552679, ..., 5.388381 , 5.388381 ,
5.388381 ],
[4.99552679, 4.99552679, 4.99552679, ..., 5.388381 , 5.388381 ,
5.388381 ],
[4.99552679, 4.99552679, 4.99552679, ..., 5.388381 , 5.388381 ,
5.388381 ],
...,
[0. , 0. , 0. , ..., 5.34579992, 5.34579992,
5.34579992],
[0. , 0. , 0. , ..., 5.34579992, 5.34579992,
5.34579992],
[0. , 0. , 0. , ..., 5.34579992, 5.34579992,
5.34579992]], shape=(75, 82)),
6: array([[5.21027994, 5.21027994, 5.21027994, ..., 5.60202217, 5.60202217,
5.60202217],
[5.21027994, 5.21027994, 5.21027994, ..., 5.60202217, 5.60202217,
5.60202217],
[5.21027994, 5.21027994, 5.21027994, ..., 5.60202217, 5.60202217,
5.60202217],
...,
[0. , 0. , 0. , ..., 5.58002138, 5.58002138,
5.58002138],
[0. , 0. , 0. , ..., 5.58002138, 5.58002138,
5.58002138],
[0. , 0. , 0. , ..., 5.58002138, 5.58002138,
5.58002138]], shape=(75, 82)),
7: array([[4.54519796, 4.54519796, 4.54519796, ..., 4.94014788, 4.94014788,
4.94014788],
[4.54519796, 4.54519796, 4.54519796, ..., 4.94014788, 4.94014788,
4.94014788],
[4.54519796, 4.54519796, 4.54519796, ..., 4.94014788, 4.94014788,
4.94014788],
...,
[0. , 0. , 0. , ..., 4.97207499, 4.97207499,
4.97207499],
[0. , 0. , 0. , ..., 4.97207499, 4.97207499,
4.97207499],
[0. , 0. , 0. , ..., 4.97207499, 4.97207499,
4.97207499]], shape=(75, 82)),
8: array([[3.38186955, 3.38186955, 3.38186955, ..., 3.73335552, 3.73335552,
3.73335552],
[3.38186955, 3.38186955, 3.38186955, ..., 3.73335552, 3.73335552,
3.73335552],
[3.38186955, 3.38186955, 3.38186955, ..., 3.73335552, 3.73335552,
3.73335552],
...,
[0. , 0. , 0. , ..., 3.75254512, 3.75254512,
3.75254512],
[0. , 0. , 0. , ..., 3.75254512, 3.75254512,
3.75254512],
[0. , 0. , 0. , ..., 3.75254512, 3.75254512,
3.75254512]], shape=(75, 82)),
9: array([[1.60437179, 1.60437179, 1.60437179, ..., 1.80872381, 1.80872381,
1.80872381],
[1.60437179, 1.60437179, 1.60437179, ..., 1.80872381, 1.80872381,
1.80872381],
[1.60437179, 1.60437179, 1.60437179, ..., 1.80872381, 1.80872381,
1.80872381],
...,
[0. , 0. , 0. , ..., 1.84118676, 1.84118676,
1.84118676],
[0. , 0. , 0. , ..., 1.84118676, 1.84118676,
1.84118676],
[0. , 0. , 0. , ..., 1.84118676, 1.84118676,
1.84118676]], shape=(75, 82)),
10: array([[1.23011148, 1.23011148, 1.23011148, ..., 1.41456532, 1.41456532,
1.41456532],
[1.23011148, 1.23011148, 1.23011148, ..., 1.41456532, 1.41456532,
1.41456532],
[1.23011148, 1.23011148, 1.23011148, ..., 1.41456532, 1.41456532,
1.41456532],
...,
[0. , 0. , 0. , ..., 1.42739856, 1.42739856,
1.42739856],
[0. , 0. , 0. , ..., 1.42739856, 1.42739856,
1.42739856],
[0. , 0. , 0. , ..., 1.42739856, 1.42739856,
1.42739856]], shape=(75, 82)),
11: array([[0.76984066, 0.76984066, 0.76984066, ..., 0.85954601, 0.85954601,
0.85954601],
[0.76984066, 0.76984066, 0.76984066, ..., 0.85954601, 0.85954601,
0.85954601],
[0.76984066, 0.76984066, 0.76984066, ..., 0.85954601, 0.85954601,
0.85954601],
...,
[0. , 0. , 0. , ..., 0.8547039 , 0.8547039 ,
0.8547039 ],
[0. , 0. , 0. , ..., 0.8547039 , 0.8547039 ,
0.8547039 ],
[0. , 0. , 0. , ..., 0.8547039 , 0.8547039 ,
0.8547039 ]], shape=(75, 82))}
And resample the precipitation raster we saved earlier
[43]:
rstr = flopy.utils.Raster.load(prcp_raster)
prcp_monthly = {}
for band in rstr.bands:
t = rstr.resample_to_grid(
modelgrid,
band,
method="nearest"
)
t[t == rstr.nodatavals[0]] = 0
prcp_monthly[band - 1] = t
prcp_monthly
[43]:
{0: array([[4.47806406, 4.47806406, 4.47806406, ..., 3.39419365, 3.39419365,
3.39419365],
[4.47806406, 4.47806406, 4.47806406, ..., 3.39419365, 3.39419365,
3.39419365],
[4.47806406, 4.47806406, 4.47806406, ..., 3.39419365, 3.39419365,
3.39419365],
...,
[0. , 0. , 0. , ..., 4.0393548 , 4.0393548 ,
4.0393548 ],
[0. , 0. , 0. , ..., 4.0393548 , 4.0393548 ,
4.0393548 ],
[0. , 0. , 0. , ..., 4.0393548 , 4.0393548 ,
4.0393548 ]], shape=(75, 82)),
1: array([[2.36392856, 2.36392856, 2.36392856, ..., 1.75142848, 1.75142848,
1.75142848],
[2.36392856, 2.36392856, 2.36392856, ..., 1.75142848, 1.75142848,
1.75142848],
[2.36392856, 2.36392856, 2.36392856, ..., 1.75142848, 1.75142848,
1.75142848],
...,
[0. , 0. , 0. , ..., 2.35964322, 2.35964322,
2.35964322],
[0. , 0. , 0. , ..., 2.35964322, 2.35964322,
2.35964322],
[0. , 0. , 0. , ..., 2.35964322, 2.35964322,
2.35964322]], shape=(75, 82)),
2: array([[2.12096763, 2.12096763, 2.12096763, ..., 1.53064525, 1.53064525,
1.53064525],
[2.12096763, 2.12096763, 2.12096763, ..., 1.53064525, 1.53064525,
1.53064525],
[2.12096763, 2.12096763, 2.12096763, ..., 1.53064525, 1.53064525,
1.53064525],
...,
[0. , 0. , 0. , ..., 1.75612891, 1.75612891,
1.75612891],
[0. , 0. , 0. , ..., 1.75612891, 1.75612891,
1.75612891],
[0. , 0. , 0. , ..., 1.75612891, 1.75612891,
1.75612891]], shape=(75, 82)),
3: array([[0.597 , 0.597 , 0.597 , ..., 0.46433336, 0.46433336,
0.46433336],
[0.597 , 0.597 , 0.597 , ..., 0.46433336, 0.46433336,
0.46433336],
[0.597 , 0.597 , 0.597 , ..., 0.46433336, 0.46433336,
0.46433336],
...,
[0. , 0. , 0. , ..., 0.53933334, 0.53933334,
0.53933334],
[0. , 0. , 0. , ..., 0.53933334, 0.53933334,
0.53933334],
[0. , 0. , 0. , ..., 0.53933334, 0.53933334,
0.53933334]], shape=(75, 82)),
4: array([[0.78161287, 0.78161287, 0.78161287, ..., 0.80645162, 0.80645162,
0.80645162],
[0.78161287, 0.78161287, 0.78161287, ..., 0.80645162, 0.80645162,
0.80645162],
[0.78161287, 0.78161287, 0.78161287, ..., 0.80645162, 0.80645162,
0.80645162],
...,
[0. , 0. , 0. , ..., 0.79193544, 0.79193544,
0.79193544],
[0. , 0. , 0. , ..., 0.79193544, 0.79193544,
0.79193544],
[0. , 0. , 0. , ..., 0.79193544, 0.79193544,
0.79193544]], shape=(75, 82)),
5: array([[0.24333334, 0.24333334, 0.24333334, ..., 0.14833333, 0.14833333,
0.14833333],
[0.24333334, 0.24333334, 0.24333334, ..., 0.14833333, 0.14833333,
0.14833333],
[0.24333334, 0.24333334, 0.24333334, ..., 0.14833333, 0.14833333,
0.14833333],
...,
[0. , 0. , 0. , ..., 0.14933333, 0.14933333,
0.14933333],
[0. , 0. , 0. , ..., 0.14933333, 0.14933333,
0.14933333],
[0. , 0. , 0. , ..., 0.14933333, 0.14933333,
0.14933333]], shape=(75, 82)),
6: array([[0.20354839, 0.20354839, 0.20354839, ..., 0.20645161, 0.20645161,
0.20645161],
[0.20354839, 0.20354839, 0.20354839, ..., 0.20645161, 0.20645161,
0.20645161],
[0.20354839, 0.20354839, 0.20354839, ..., 0.20645161, 0.20645161,
0.20645161],
...,
[0. , 0. , 0. , ..., 0.20451613, 0.20451613,
0.20451613],
[0. , 0. , 0. , ..., 0.20451613, 0.20451613,
0.20451613],
[0. , 0. , 0. , ..., 0.20451613, 0.20451613,
0.20451613]], shape=(75, 82)),
7: array([[0.08483871, 0.08483871, 0.08483871, ..., 0.11064516, 0.11064516,
0.11064516],
[0.08483871, 0.08483871, 0.08483871, ..., 0.11064516, 0.11064516,
0.11064516],
[0.08483871, 0.08483871, 0.08483871, ..., 0.11064516, 0.11064516,
0.11064516],
...,
[0. , 0. , 0. , ..., 0. , 0. ,
0. ],
[0. , 0. , 0. , ..., 0. , 0. ,
0. ],
[0. , 0. , 0. , ..., 0. , 0. ,
0. ]], shape=(75, 82)),
8: array([[0.30833334, 0.30833334, 0.30833334, ..., 0.22766666, 0.22766666,
0.22766666],
[0.30833334, 0.30833334, 0.30833334, ..., 0.22766666, 0.22766666,
0.22766666],
[0.30833334, 0.30833334, 0.30833334, ..., 0.22766666, 0.22766666,
0.22766666],
...,
[0. , 0. , 0. , ..., 0.48166665, 0.48166665,
0.48166665],
[0. , 0. , 0. , ..., 0.48166665, 0.48166665,
0.48166665],
[0. , 0. , 0. , ..., 0.48166665, 0.48166665,
0.48166665]], shape=(75, 82)),
9: array([[9.75677395, 9.75677395, 9.75677395, ..., 8.6554842 , 8.6554842 ,
8.6554842 ],
[9.75677395, 9.75677395, 9.75677395, ..., 8.6554842 , 8.6554842 ,
8.6554842 ],
[9.75677395, 9.75677395, 9.75677395, ..., 8.6554842 , 8.6554842 ,
8.6554842 ],
...,
[0. , 0. , 0. , ..., 8.43290234, 8.43290234,
8.43290234],
[0. , 0. , 0. , ..., 8.43290234, 8.43290234,
8.43290234],
[0. , 0. , 0. , ..., 8.43290234, 8.43290234,
8.43290234]], shape=(75, 82)),
10: array([[1.15666664, 1.15666664, 1.15666664, ..., 0.90833336, 0.90833336,
0.90833336],
[1.15666664, 1.15666664, 1.15666664, ..., 0.90833336, 0.90833336,
0.90833336],
[1.15666664, 1.15666664, 1.15666664, ..., 0.90833336, 0.90833336,
0.90833336],
...,
[0. , 0. , 0. , ..., 1.10366654, 1.10366654,
1.10366654],
[0. , 0. , 0. , ..., 1.10366654, 1.10366654,
1.10366654],
[0. , 0. , 0. , ..., 1.10366654, 1.10366654,
1.10366654]], shape=(75, 82)),
11: array([[11.07774162, 11.07774162, 11.07774162, ..., 9.146451 ,
9.146451 , 9.146451 ],
[11.07774162, 11.07774162, 11.07774162, ..., 9.146451 ,
9.146451 , 9.146451 ],
[11.07774162, 11.07774162, 11.07774162, ..., 9.146451 ,
9.146451 , 9.146451 ],
...,
[ 0. , 0. , 0. , ..., 10.41838837,
10.41838837, 10.41838837],
[ 0. , 0. , 0. , ..., 10.41838837,
10.41838837, 10.41838837],
[ 0. , 0. , 0. , ..., 10.41838837,
10.41838837, 10.41838837]], shape=(75, 82))}
Potetial evapotranspiration and Precipitation data from daymet are in mm/day, however our discretization is meters/day.
[44]:
prcp_monthly = {k: v * 0.001 for k, v in prcp_monthly.items()}
pet_monthly = {k: v * 0.001 for k, v in pet_monthly.items()}
Finally we can load a saved soil hydraulic conductivity array, and we have the information we need to create a Unsaturated Zone Flow (UZF) package for this model
[45]:
ksat_raster = data_path / "ksat.img"
rstr = flopy.utils.Raster.load(ksat_raster)
vks = rstr.resample_to_grid(
modelgrid,
rstr.bands[0],
method="nearest"
)
vks[vks == rstr.nodatavals[0]] = np.nan
vks[np.isnan(vks)] = np.nanmean(vks)
vks *= 0.0864
ims = plt.imshow(vks)
plt.colorbar(shrink=0.6); # um per sec to m per day
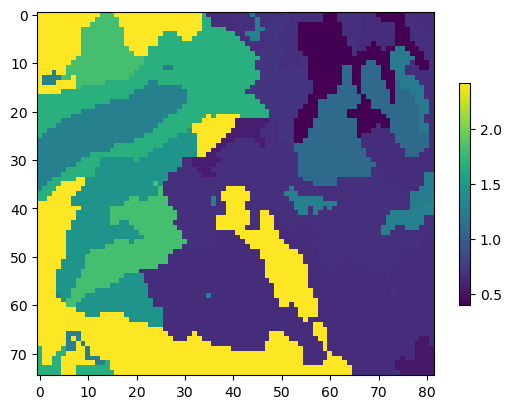
Creating a UZF package
Now that we have some climate and soils data we can create a UZF package from it and add it to the model!
Create the package data and the stress period data info for flopy
[46]:
modelgrid = gwf.modelgrid
modelgrid.set_coord_info(xoff=xmin, yoff=ymin)
package_data = []
cnt = 0
for i in range(modelgrid.nrow):
for j in range(modelgrid.ncol):
if modelgrid.idomain[0, i, j] == 0:
continue
rec = (cnt, (0, i, j), 1, 0, 1.0, vks[i, j], 0.1, 0.38, 0.1, 3.5)
package_data.append(rec)
cnt += 1
period_data = {}
for per in range(gwf.nper):
spd = []
cnt = 0
for i in range(modelgrid.nrow * modelgrid.ncol):
if modelgrid.idomain.ravel()[i] == 0:
continue
rec = (cnt, prcp_monthly[per].ravel()[i], pet_monthly[per].ravel()[i], 0.5, 0.2, -1.1, -75, 1.0)
spd.append(rec)
cnt += 1
period_data[per] = spd
[47]:
uzf = flopy.mf6.ModflowGwfuzf(
gwf,
simulate_et=True,
ntrailwaves=15,
nwavesets=100,
packagedata=package_data,
perioddata=period_data,
unsat_etwc=True,
linear_gwet=True,
simulate_gwseep=True
)
Now to create a simple river package from NHD flowlines
We can use the intersection routines that have been presented thus far to create a river package for the sagehen model.
The first step is load the NHD flowlines for the basin
[48]:
flow_lines = data_path / "sagehen_nhd.shp"
flw_all = gpd.read_file(flow_lines)
flw_all.plot();
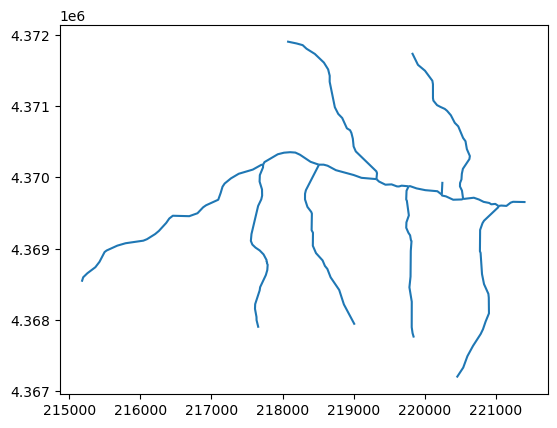
Excellent! Now we have all of the flow lines for the Sagehen watershed. But what if we want to identify the main stem?
[49]:
flow_lines = data_path / "sagehen_main_nhd.shp"
flw_main = gpd.read_file(flow_lines)
flw_main.plot()
flw_main
[49]:
| nhdplus_co | geometry | |
|---|---|---|
| 0 | 8933522 | LINESTRING (221038.128 4369598.916, 221076.482... |
| 1 | 8933524 | LINESTRING (220540.741 4369697.577, 220691.351... |
| 2 | 8933520 | LINESTRING (220240.365 4369758.86, 220244.995 ... |
| 3 | 8934344 | LINESTRING (219769.843 4369876.335, 219795.329... |
| 4 | 8933512 | LINESTRING (219321.052 4369977.09, 219357.89 4... |
| 5 | 8933508 | LINESTRING (218509.989 4370180.464, 218578.637... |
| 6 | 8933496 | LINESTRING (217726.838 4370179.373, 217746.044... |
| 7 | 8933582 | LINESTRING (215175.791 4368533.473, 215195.962... |
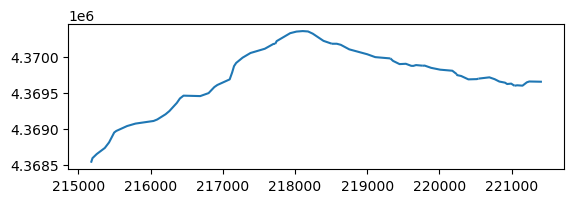
Great! Now we can start processing data to create a river package. We can first identify our main stem cells.
[50]:
main_stem = []
gx = GridIntersect(modelgrid)
for iloc, row in flw_main.iterrows():
results = gx.intersect(row.geometry)
main_stem += list(results.cellids)
main_stem[0:5]
/Users/mnfienen/miniforge3/envs/pyclass/lib/python3.11/site-packages/flopy/utils/gridintersect.py:123: DeprecationWarning: Note `method="structured"` is deprecated. Pass `method="vertex"` to silence this warning. This will be the new default in a future release and this keyword argument will be removed.
warnings.warn(
[50]:
[(42, 75), (42, 76), (41, 76), (41, 77), (41, 78)]
Class Exercise:
Use GridIntersect to create a list of tributary cells from the flw_all geodataframe.
Hint Use the main_stem list to filter out cells that are associated with the main stem of the river and only store the cells that are associated with tributaries to the main stem.
[51]:
tributaries = []
gx = GridIntersect(modelgrid)
for iloc, row in flw_all.iterrows():
results = gx.intersect(row.geometry)
for res in results.cellids:
if res in main_stem:
continue
else:
tributaries.append(res)
tributaries[0:5]
with open(data_path / "trib_cells.txt", 'w') as foo:
for line in tributaries:
foo.write(f"{line[0]},{line[1]}\n")
/Users/mnfienen/miniforge3/envs/pyclass/lib/python3.11/site-packages/flopy/utils/gridintersect.py:123: DeprecationWarning: Note `method="structured"` is deprecated. Pass `method="vertex"` to silence this warning. This will be the new default in a future release and this keyword argument will be removed.
warnings.warn(
The cellids for tributaries have been stored previously, so we’ll load those up and continue on.
[52]:
tributaries = []
with open(data_path / "trib_cells.txt") as foo:
for line in foo:
t = line.strip().split(",")
tributaries.append((int(t[0]), int(t[1])))
We can also get daily discharge data to help calculate average monthly stages for the RIV package
We’ll use the dataretrieval package to get stage information for our model
[53]:
import dataretrieval.nwis as nwis
gage = nwis.get_record(sites=station_id, service='iv', start="2023-01-01", end="2023-12-31", parameterCd="00065")
gage.to_csv(data_path / "sagehen_gage_data.csv")
[54]:
gage = pd.read_csv(data_path / "sagehen_gage_data.csv")
gage["datetime"] = pd.to_datetime(gage["datetime"])
gage = gage.set_index("datetime")
gage.rename(columns={"00065": "stage"}, inplace=True)
gage.stage *= 0.3284
ax = gage.stage.plot()
ax.set_ylabel("stage, in feet")
gage.head()
[54]:
| site_no | stage | 00065_cd | |
|---|---|---|---|
| datetime | |||
| 2023-01-01 08:00:00+00:00 | 10343500 | 0.748752 | A |
| 2023-01-01 08:15:00+00:00 | 10343500 | 0.748752 | A |
| 2023-01-01 08:30:00+00:00 | 10343500 | 0.748752 | A |
| 2023-01-01 08:45:00+00:00 | 10343500 | 0.748752 | A |
| 2023-01-01 09:00:00+00:00 | 10343500 | 0.748752 | A |
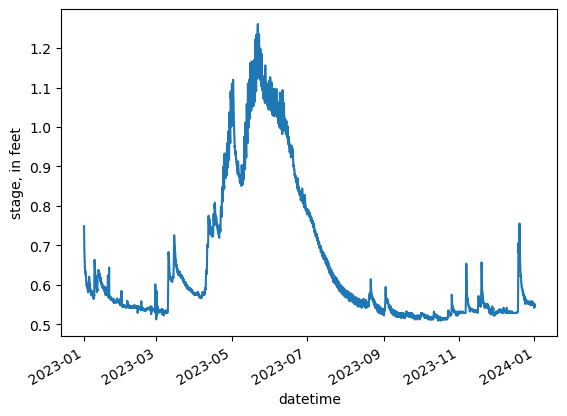
Now we can aggregate to a mean monthly stage from 15 minute instantaneous values
[55]:
gage["month"] = gage.index.month
gage_mmo = gage.groupby(by=["month"], as_index=False)["stage"].mean()
ax = gage_mmo.stage.plot()
ax.set_ylabel("mean stage, in feet");
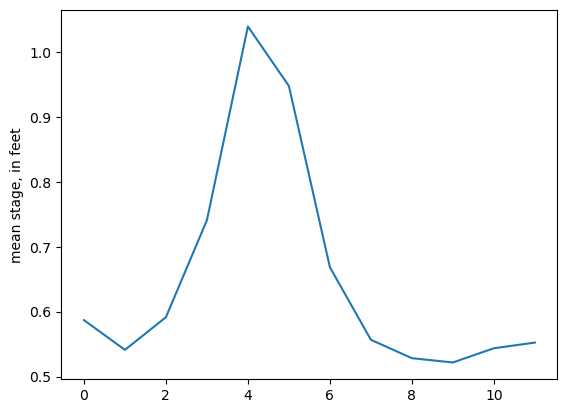
Now to create the RIV package
[56]:
cond = 10
rbadj = 1
stage_adj = 0.33
perioddata = {}
for kper, row in gage_mmo.iterrows():
spd = []
for (i, j) in main_stem:
ctop = modelgrid.top[i, j]
rbed = ctop - rbadj
rec = ((0, i, j), row.stage + rbed, cond, rbed)
spd.append(rec)
for (i, j) in tributaries:
ctop = modelgrid.top[i, j]
rbed = ctop - (rbadj / 2)
rec = ((0, i, j), row.stage + rbed, cond, rbed)
perioddata[kper] = spd
perioddata[0][0:5]
[56]:
[((0, 42, 75),
np.float64(1936.8217418655074),
10,
np.float64(1936.2347412109375)),
((0, 42, 76), np.float64(1935.65633659207), 10, np.float64(1935.0693359375)),
((0, 41, 76),
np.float64(1932.400721357695),
10,
np.float64(1931.813720703125)),
((0, 41, 77),
np.float64(1930.8251598342574),
10,
np.float64(1930.2381591796875)),
((0, 41, 78), np.float64(1928.34823112332), 10, np.float64(1927.76123046875))]
[57]:
riv = flopy.mf6.ModflowGwfriv(
gwf,
save_flows=True,
maxbound=len(perioddata[0]),
stress_period_data=perioddata
)
Testing the model
[58]:
sim.write_simulation()
sim.run_simulation();
writing simulation...
writing simulation name file...
writing simulation tdis package...
writing solution package ims_-1...
writing model sagehen...
writing model name file...
writing package dis...
writing package ic...
writing package npf...
writing package sto...
writing package oc...
writing package uzf_0...
INFORMATION: nuzfcells in ('gwf6', 'uzf', 'dimensions') changed to 3587 based on size of packagedata
writing package riv_0...
FloPy is using the following executable to run the model: ../../../../../../../.local/share/flopy/bin/mf6
MODFLOW 6
U.S. GEOLOGICAL SURVEY MODULAR HYDROLOGIC MODEL
VERSION 6.4.2 06/28/2023
MODFLOW 6 compiled Jul 05 2023 20:29:14 with Intel(R) Fortran Intel(R) 64
Compiler Classic for applications running on Intel(R) 64, Version 2021.7.0
Build 20220726_000000
This software has been approved for release by the U.S. Geological
Survey (USGS). Although the software has been subjected to rigorous
review, the USGS reserves the right to update the software as needed
pursuant to further analysis and review. No warranty, expressed or
implied, is made by the USGS or the U.S. Government as to the
functionality of the software and related material nor shall the
fact of release constitute any such warranty. Furthermore, the
software is released on condition that neither the USGS nor the U.S.
Government shall be held liable for any damages resulting from its
authorized or unauthorized use. Also refer to the USGS Water
Resources Software User Rights Notice for complete use, copyright,
and distribution information.
Run start date and time (yyyy/mm/dd hh:mm:ss): 2025/04/27 10:59:44
Writing simulation list file: mfsim.lst
Using Simulation name file: mfsim.nam
Solving: Stress period: 1 Time step: 1
Solving: Stress period: 1 Time step: 2
Solving: Stress period: 1 Time step: 3
Solving: Stress period: 1 Time step: 4
Solving: Stress period: 1 Time step: 5
Solving: Stress period: 1 Time step: 6
Solving: Stress period: 1 Time step: 7
Solving: Stress period: 1 Time step: 8
Solving: Stress period: 1 Time step: 9
Solving: Stress period: 1 Time step: 10
Solving: Stress period: 1 Time step: 11
Solving: Stress period: 1 Time step: 12
Solving: Stress period: 1 Time step: 13
Solving: Stress period: 1 Time step: 14
Solving: Stress period: 1 Time step: 15
Solving: Stress period: 1 Time step: 16
Solving: Stress period: 1 Time step: 17
Solving: Stress period: 1 Time step: 18
Solving: Stress period: 1 Time step: 19
Solving: Stress period: 1 Time step: 20
Solving: Stress period: 1 Time step: 21
Solving: Stress period: 1 Time step: 22
Solving: Stress period: 1 Time step: 23
Solving: Stress period: 1 Time step: 24
Solving: Stress period: 1 Time step: 25
Solving: Stress period: 1 Time step: 26
Solving: Stress period: 1 Time step: 27
Solving: Stress period: 1 Time step: 28
Solving: Stress period: 1 Time step: 29
Solving: Stress period: 1 Time step: 30
Solving: Stress period: 1 Time step: 31
Solving: Stress period: 2 Time step: 1
Solving: Stress period: 2 Time step: 2
Solving: Stress period: 2 Time step: 3
Solving: Stress period: 2 Time step: 4
Solving: Stress period: 2 Time step: 5
Solving: Stress period: 2 Time step: 6
Solving: Stress period: 2 Time step: 7
Solving: Stress period: 2 Time step: 8
Solving: Stress period: 2 Time step: 9
Solving: Stress period: 2 Time step: 10
Solving: Stress period: 2 Time step: 11
Solving: Stress period: 2 Time step: 12
Solving: Stress period: 2 Time step: 13
Solving: Stress period: 2 Time step: 14
Solving: Stress period: 2 Time step: 15
Solving: Stress period: 2 Time step: 16
Solving: Stress period: 2 Time step: 17
Solving: Stress period: 2 Time step: 18
Solving: Stress period: 2 Time step: 19
Solving: Stress period: 2 Time step: 20
Solving: Stress period: 2 Time step: 21
Solving: Stress period: 2 Time step: 22
Solving: Stress period: 2 Time step: 23
Solving: Stress period: 2 Time step: 24
Solving: Stress period: 2 Time step: 25
Solving: Stress period: 2 Time step: 26
Solving: Stress period: 2 Time step: 27
Solving: Stress period: 2 Time step: 28
Solving: Stress period: 3 Time step: 1
Solving: Stress period: 3 Time step: 2
Solving: Stress period: 3 Time step: 3
Solving: Stress period: 3 Time step: 4
Solving: Stress period: 3 Time step: 5
Solving: Stress period: 3 Time step: 6
Solving: Stress period: 3 Time step: 7
Solving: Stress period: 3 Time step: 8
Solving: Stress period: 3 Time step: 9
Solving: Stress period: 3 Time step: 10
Solving: Stress period: 3 Time step: 11
Solving: Stress period: 3 Time step: 12
Solving: Stress period: 3 Time step: 13
Solving: Stress period: 3 Time step: 14
Solving: Stress period: 3 Time step: 15
Solving: Stress period: 3 Time step: 16
Solving: Stress period: 3 Time step: 17
Solving: Stress period: 3 Time step: 18
Solving: Stress period: 3 Time step: 19
Solving: Stress period: 3 Time step: 20
Solving: Stress period: 3 Time step: 21
Solving: Stress period: 3 Time step: 22
Solving: Stress period: 3 Time step: 23
Solving: Stress period: 3 Time step: 24
Solving: Stress period: 3 Time step: 25
Solving: Stress period: 3 Time step: 26
Solving: Stress period: 3 Time step: 27
Solving: Stress period: 3 Time step: 28
Solving: Stress period: 3 Time step: 29
Solving: Stress period: 3 Time step: 30
Solving: Stress period: 3 Time step: 31
Solving: Stress period: 4 Time step: 1
Solving: Stress period: 4 Time step: 2
Solving: Stress period: 4 Time step: 3
Solving: Stress period: 4 Time step: 4
Solving: Stress period: 4 Time step: 5
Solving: Stress period: 4 Time step: 6
Solving: Stress period: 4 Time step: 7
Solving: Stress period: 4 Time step: 8
Solving: Stress period: 4 Time step: 9
Solving: Stress period: 4 Time step: 10
Solving: Stress period: 4 Time step: 11
Solving: Stress period: 4 Time step: 12
Solving: Stress period: 4 Time step: 13
Solving: Stress period: 4 Time step: 14
Solving: Stress period: 4 Time step: 15
Solving: Stress period: 4 Time step: 16
Solving: Stress period: 4 Time step: 17
Solving: Stress period: 4 Time step: 18
Solving: Stress period: 4 Time step: 19
Solving: Stress period: 4 Time step: 20
Solving: Stress period: 4 Time step: 21
Solving: Stress period: 4 Time step: 22
Solving: Stress period: 4 Time step: 23
Solving: Stress period: 4 Time step: 24
Solving: Stress period: 4 Time step: 25
Solving: Stress period: 4 Time step: 26
Solving: Stress period: 4 Time step: 27
Solving: Stress period: 4 Time step: 28
Solving: Stress period: 4 Time step: 29
Solving: Stress period: 4 Time step: 30
Solving: Stress period: 5 Time step: 1
Solving: Stress period: 5 Time step: 2
Solving: Stress period: 5 Time step: 3
Solving: Stress period: 5 Time step: 4
Solving: Stress period: 5 Time step: 5
Solving: Stress period: 5 Time step: 6
Solving: Stress period: 5 Time step: 7
Solving: Stress period: 5 Time step: 8
Solving: Stress period: 5 Time step: 9
Solving: Stress period: 5 Time step: 10
Solving: Stress period: 5 Time step: 11
Solving: Stress period: 5 Time step: 12
Solving: Stress period: 5 Time step: 13
Solving: Stress period: 5 Time step: 14
Solving: Stress period: 5 Time step: 15
Solving: Stress period: 5 Time step: 16
Solving: Stress period: 5 Time step: 17
Solving: Stress period: 5 Time step: 18
Solving: Stress period: 5 Time step: 19
Solving: Stress period: 5 Time step: 20
Solving: Stress period: 5 Time step: 21
Solving: Stress period: 5 Time step: 22
Solving: Stress period: 5 Time step: 23
Solving: Stress period: 5 Time step: 24
Solving: Stress period: 5 Time step: 25
Solving: Stress period: 5 Time step: 26
Solving: Stress period: 5 Time step: 27
Solving: Stress period: 5 Time step: 28
Solving: Stress period: 5 Time step: 29
Solving: Stress period: 5 Time step: 30
Solving: Stress period: 5 Time step: 31
Solving: Stress period: 6 Time step: 1
Solving: Stress period: 6 Time step: 2
Solving: Stress period: 6 Time step: 3
Solving: Stress period: 6 Time step: 4
Solving: Stress period: 6 Time step: 5
Solving: Stress period: 6 Time step: 6
Solving: Stress period: 6 Time step: 7
Solving: Stress period: 6 Time step: 8
Solving: Stress period: 6 Time step: 9
Solving: Stress period: 6 Time step: 10
Solving: Stress period: 6 Time step: 11
Solving: Stress period: 6 Time step: 12
Solving: Stress period: 6 Time step: 13
Solving: Stress period: 6 Time step: 14
Solving: Stress period: 6 Time step: 15
Solving: Stress period: 6 Time step: 16
Solving: Stress period: 6 Time step: 17
Solving: Stress period: 6 Time step: 18
Solving: Stress period: 6 Time step: 19
Solving: Stress period: 6 Time step: 20
Solving: Stress period: 6 Time step: 21
Solving: Stress period: 6 Time step: 22
Solving: Stress period: 6 Time step: 23
Solving: Stress period: 6 Time step: 24
Solving: Stress period: 6 Time step: 25
Solving: Stress period: 6 Time step: 26
Solving: Stress period: 6 Time step: 27
Solving: Stress period: 6 Time step: 28
Solving: Stress period: 6 Time step: 29
Solving: Stress period: 6 Time step: 30
Solving: Stress period: 7 Time step: 1
Solving: Stress period: 7 Time step: 2
Solving: Stress period: 7 Time step: 3
Solving: Stress period: 7 Time step: 4
Solving: Stress period: 7 Time step: 5
Solving: Stress period: 7 Time step: 6
Solving: Stress period: 7 Time step: 7
Solving: Stress period: 7 Time step: 8
Solving: Stress period: 7 Time step: 9
Solving: Stress period: 7 Time step: 10
Solving: Stress period: 7 Time step: 11
Solving: Stress period: 7 Time step: 12
Solving: Stress period: 7 Time step: 13
Solving: Stress period: 7 Time step: 14
Solving: Stress period: 7 Time step: 15
Solving: Stress period: 7 Time step: 16
Solving: Stress period: 7 Time step: 17
Solving: Stress period: 7 Time step: 18
Solving: Stress period: 7 Time step: 19
Solving: Stress period: 7 Time step: 20
Solving: Stress period: 7 Time step: 21
Solving: Stress period: 7 Time step: 22
Solving: Stress period: 7 Time step: 23
Solving: Stress period: 7 Time step: 24
Solving: Stress period: 7 Time step: 25
Solving: Stress period: 7 Time step: 26
Solving: Stress period: 7 Time step: 27
Solving: Stress period: 7 Time step: 28
Solving: Stress period: 7 Time step: 29
Solving: Stress period: 7 Time step: 30
Solving: Stress period: 7 Time step: 31
Solving: Stress period: 8 Time step: 1
Solving: Stress period: 8 Time step: 2
Solving: Stress period: 8 Time step: 3
Solving: Stress period: 8 Time step: 4
Solving: Stress period: 8 Time step: 5
Solving: Stress period: 8 Time step: 6
Solving: Stress period: 8 Time step: 7
Solving: Stress period: 8 Time step: 8
Solving: Stress period: 8 Time step: 9
Solving: Stress period: 8 Time step: 10
Solving: Stress period: 8 Time step: 11
Solving: Stress period: 8 Time step: 12
Solving: Stress period: 8 Time step: 13
Solving: Stress period: 8 Time step: 14
Solving: Stress period: 8 Time step: 15
Solving: Stress period: 8 Time step: 16
Solving: Stress period: 8 Time step: 17
Solving: Stress period: 8 Time step: 18
Solving: Stress period: 8 Time step: 19
Solving: Stress period: 8 Time step: 20
Solving: Stress period: 8 Time step: 21
Solving: Stress period: 8 Time step: 22
Solving: Stress period: 8 Time step: 23
Solving: Stress period: 8 Time step: 24
Solving: Stress period: 8 Time step: 25
Solving: Stress period: 8 Time step: 26
Solving: Stress period: 8 Time step: 27
Solving: Stress period: 8 Time step: 28
Solving: Stress period: 8 Time step: 29
Solving: Stress period: 8 Time step: 30
Solving: Stress period: 8 Time step: 31
Solving: Stress period: 9 Time step: 1
Solving: Stress period: 9 Time step: 2
Solving: Stress period: 9 Time step: 3
Solving: Stress period: 9 Time step: 4
Solving: Stress period: 9 Time step: 5
Solving: Stress period: 9 Time step: 6
Solving: Stress period: 9 Time step: 7
Solving: Stress period: 9 Time step: 8
Solving: Stress period: 9 Time step: 9
Solving: Stress period: 9 Time step: 10
Solving: Stress period: 9 Time step: 11
Solving: Stress period: 9 Time step: 12
Solving: Stress period: 9 Time step: 13
Solving: Stress period: 9 Time step: 14
Solving: Stress period: 9 Time step: 15
Solving: Stress period: 9 Time step: 16
Solving: Stress period: 9 Time step: 17
Solving: Stress period: 9 Time step: 18
Solving: Stress period: 9 Time step: 19
Solving: Stress period: 9 Time step: 20
Solving: Stress period: 9 Time step: 21
Solving: Stress period: 9 Time step: 22
Solving: Stress period: 9 Time step: 23
Solving: Stress period: 9 Time step: 24
Solving: Stress period: 9 Time step: 25
Solving: Stress period: 9 Time step: 26
Solving: Stress period: 9 Time step: 27
Solving: Stress period: 9 Time step: 28
Solving: Stress period: 9 Time step: 29
Solving: Stress period: 9 Time step: 30
Solving: Stress period: 10 Time step: 1
Solving: Stress period: 10 Time step: 2
Solving: Stress period: 10 Time step: 3
Solving: Stress period: 10 Time step: 4
Solving: Stress period: 10 Time step: 5
Solving: Stress period: 10 Time step: 6
Solving: Stress period: 10 Time step: 7
Solving: Stress period: 10 Time step: 8
Solving: Stress period: 10 Time step: 9
Solving: Stress period: 10 Time step: 10
Solving: Stress period: 10 Time step: 11
Solving: Stress period: 10 Time step: 12
Solving: Stress period: 10 Time step: 13
Solving: Stress period: 10 Time step: 14
Solving: Stress period: 10 Time step: 15
Solving: Stress period: 10 Time step: 16
Solving: Stress period: 10 Time step: 17
Solving: Stress period: 10 Time step: 18
Solving: Stress period: 10 Time step: 19
Solving: Stress period: 10 Time step: 20
Solving: Stress period: 10 Time step: 21
Solving: Stress period: 10 Time step: 22
Solving: Stress period: 10 Time step: 23
Solving: Stress period: 10 Time step: 24
Solving: Stress period: 10 Time step: 25
Solving: Stress period: 10 Time step: 26
Solving: Stress period: 10 Time step: 27
Solving: Stress period: 10 Time step: 28
Solving: Stress period: 10 Time step: 29
Solving: Stress period: 10 Time step: 30
Solving: Stress period: 10 Time step: 31
Solving: Stress period: 11 Time step: 1
Solving: Stress period: 11 Time step: 2
Solving: Stress period: 11 Time step: 3
Solving: Stress period: 11 Time step: 4
Solving: Stress period: 11 Time step: 5
Solving: Stress period: 11 Time step: 6
Solving: Stress period: 11 Time step: 7
Solving: Stress period: 11 Time step: 8
Solving: Stress period: 11 Time step: 9
Solving: Stress period: 11 Time step: 10
Solving: Stress period: 11 Time step: 11
Solving: Stress period: 11 Time step: 12
Solving: Stress period: 11 Time step: 13
Solving: Stress period: 11 Time step: 14
Solving: Stress period: 11 Time step: 15
Solving: Stress period: 11 Time step: 16
Solving: Stress period: 11 Time step: 17
Solving: Stress period: 11 Time step: 18
Solving: Stress period: 11 Time step: 19
Solving: Stress period: 11 Time step: 20
Solving: Stress period: 11 Time step: 21
Solving: Stress period: 11 Time step: 22
Solving: Stress period: 11 Time step: 23
Solving: Stress period: 11 Time step: 24
Solving: Stress period: 11 Time step: 25
Solving: Stress period: 11 Time step: 26
Solving: Stress period: 11 Time step: 27
Solving: Stress period: 11 Time step: 28
Solving: Stress period: 11 Time step: 29
Solving: Stress period: 11 Time step: 30
Solving: Stress period: 12 Time step: 1
Solving: Stress period: 12 Time step: 2
Solving: Stress period: 12 Time step: 3
Solving: Stress period: 12 Time step: 4
Solving: Stress period: 12 Time step: 5
Solving: Stress period: 12 Time step: 6
Solving: Stress period: 12 Time step: 7
Solving: Stress period: 12 Time step: 8
Solving: Stress period: 12 Time step: 9
Solving: Stress period: 12 Time step: 10
Solving: Stress period: 12 Time step: 11
Solving: Stress period: 12 Time step: 12
Solving: Stress period: 12 Time step: 13
Solving: Stress period: 12 Time step: 14
Solving: Stress period: 12 Time step: 15
Solving: Stress period: 12 Time step: 16
Solving: Stress period: 12 Time step: 17
Solving: Stress period: 12 Time step: 18
Solving: Stress period: 12 Time step: 19
Solving: Stress period: 12 Time step: 20
Solving: Stress period: 12 Time step: 21
Solving: Stress period: 12 Time step: 22
Solving: Stress period: 12 Time step: 23
Solving: Stress period: 12 Time step: 24
Solving: Stress period: 12 Time step: 25
Solving: Stress period: 12 Time step: 26
Solving: Stress period: 12 Time step: 27
Solving: Stress period: 12 Time step: 28
Solving: Stress period: 12 Time step: 29
Solving: Stress period: 12 Time step: 30
Solving: Stress period: 12 Time step: 31
Run end date and time (yyyy/mm/dd hh:mm:ss): 2025/04/27 11:00:56
Elapsed run time: 1 Minutes, 11.569 Seconds
Normal termination of simulation.
[59]:
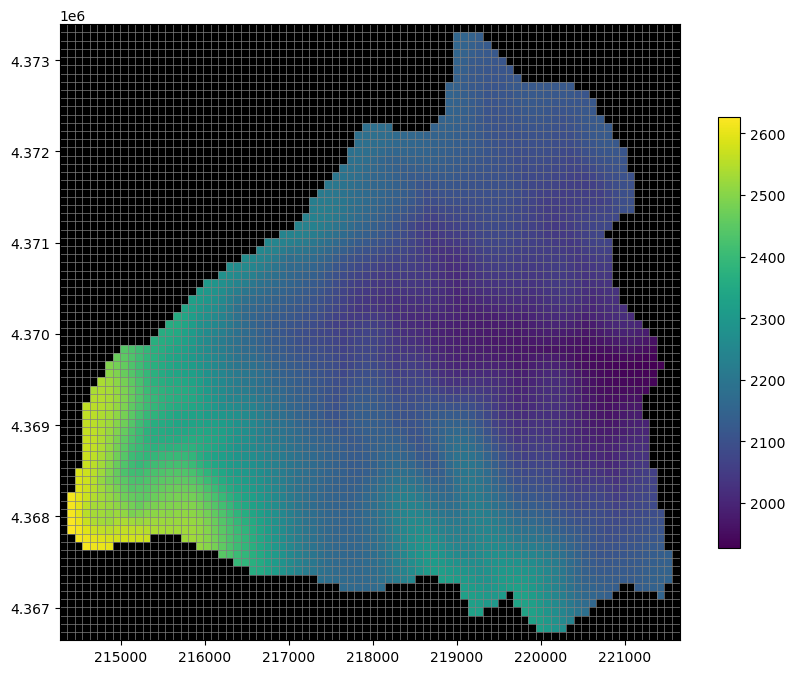
hds = gwf.output.head()
heads = hds.get_alldata()[-1]
fig, ax = plt.subplots(figsize=(10, 8))
pmv = flopy.plot.PlotMapView(modelgrid=modelgrid, ax=ax)
pc = pmv.plot_array(heads, cmap='viridis', masked_values=[1e30, -1e30])
pmv.plot_grid(lw=0.5)
pmv.plot_inactive()
plt.colorbar(pc, shrink=0.7)
plt.show();
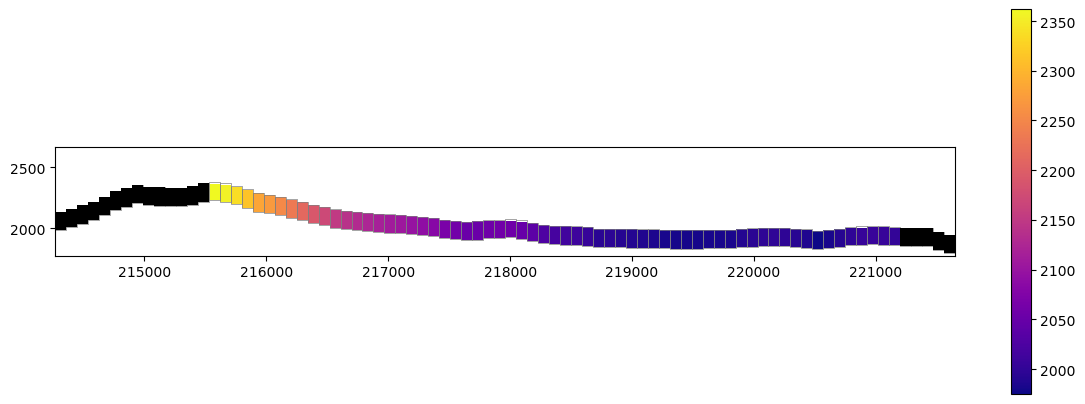
[60]:
fig, ax = plt.subplots(figsize=(12, 8))
ax.set_aspect("equal")
xc = flopy.plot.PlotCrossSection(modelgrid=modelgrid, ax=ax, line={"row": 36}, geographic_coords=True)
pc = xc.plot_array(heads, cmap='plasma', head=heads, masked_values=[1e+30])
xc.plot_grid(lw=0.5)
xc.plot_inactive()
plt.colorbar(pc, shrink=0.50)
plt.tight_layout();
[ ]: