05: Unstructured grid and general mesh generation with FloPy
In this notebook we’ll cover how to build Structured and Unstructured Meshes (Grids) with FloPy for use in MODFLOW models.
This notebook will start with generating a simple Structured Grid and we will use this grid object to begin developing Unstructured Meshes. The grids generated in theis notebook include the following:
Regular structured grid (review)
Irregular structured grid with variable row and column spacings
Local Grid Refinement (LGR): A locally refined mesh that can be subsetted into a second model within a MODFLOW simulation
Quadtree Mesh: A mesh with a four fold (quad) refinement for each grid cell in a given refinement area
Triangular Mesh: A triangular mesh generated using Delaunay triangulation
Voronoi Mesh: A mesh generated from the connected centers of the Delaunay triangulation circumcircles
[1]:
import warnings
from pathlib import Path
import flopy
import geopandas as gpd
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import contextily as ctx
from dataretrieval import nldi, nwis
from flopy.discretization import StructuredGrid, VertexGrid
from flopy.plot import PlotMapView
from flopy.utils.gridgen import Gridgen
from flopy.utils.gridintersect import GridIntersect
from flopy.utils.lgrutil import Lgr
from flopy.utils.rasters import Raster
from flopy.utils.triangle import Triangle
from flopy.utils.voronoi import VoronoiGrid
from shapely.errors import ShapelyDeprecationWarning
warnings.simplefilter("ignore", DeprecationWarning)
warnings.simplefilter("ignore", ShapelyDeprecationWarning)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[1], line 9
7 import numpy as np
8 import pandas as pd
----> 9 import contextily as ctx
10 from dataretrieval import nldi, nwis
11 from flopy.discretization import StructuredGrid, VertexGrid
ModuleNotFoundError: No module named 'contextily'
Creating a regular structured (rectilinear) grid
This activity will start by creating a simple sturctured (rectilinear) grid from a 1/3 arc second Digital Elevation Model of the Sagehen Creek watershed as an illustration of how we can create an initial mesh that can be used to develop a model from.
This is a quick review for more information, see the previous activity 04_Modelgrid_and_intersection.ipynb
Load the Digital Elevation Model and visualize it
[2]:
data_ws = Path("./data")
geospatial_ws = data_ws / "unstructured_grid_data"
dem = geospatial_ws / "dem.img"
rstr = Raster.load(dem)
rstr.plot();
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[2], line 4
2 geospatial_ws = data_ws / "unstructured_grid_data"
3 dem = geospatial_ws / "dem.img"
----> 4 rstr = Raster.load(dem)
5 rstr.plot();
NameError: name 'Raster' is not defined
Get the DEM boundaries
[3]:
xmin, xmax, ymin, ymax = rstr.bounds
epsg = rstr.crs.to_epsg()
print(f"{xmin=}, {xmax=}")
print(f"{epsg=}")
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[3], line 1
----> 1 xmin, xmax, ymin, ymax = rstr.bounds
2 epsg = rstr.crs.to_epsg()
3 print(f"{xmin=}, {xmax=}")
NameError: name 'rstr' is not defined
And begin developing an initial mesh for the model
[4]:
# live code modelgrid creation
cellsize = 200
Finally create a fake top, bottom, and idomain for the model for now. These will be refined in a subsequent step.
[5]:
# live code modelgrid creation
And now we can generate a StructuredGrid object (called sgrid) that can be used for developing model inputs
[6]:
# live code modelgrid creation
We can plot our SturcturedGrid object over the existing raster data to check that it is correctly orientied in space
[7]:
fig, ax = plt.subplots(figsize=(6, 6))
rstr.plot(ax=ax)
pmv = PlotMapView(modelgrid=sgrid, ax=ax)
pmv.plot_grid(lw=0.5);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[7], line 2
1 fig, ax = plt.subplots(figsize=(6, 6))
----> 2 rstr.plot(ax=ax)
3 pmv = PlotMapView(modelgrid=sgrid, ax=ax)
4 pmv.plot_grid(lw=0.5);
NameError: name 'rstr' is not defined
Resampling data to finish creating our initial grid.
Once we have an initial grid, raster and vector data can be resampled or intersected with the grid to begin building a model domain. Although the focus of this notebook is to generate different types of meshes for modeling, the resampling and intersection process in this notebook can be applied many different data types and be used to generate boundary conditions within a MODFLOW model.
Raster resampling
We’ll use FloPy’s built in raster resampling method to generate values for land surface elevation (top) using the existing StructuredGrid object. For more information on raster resampling, please see this example
[8]:
top = rstr.resample_to_grid(
sgrid, rstr.bands[0], method="min", extrapolate_edges=True
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[8], line 1
----> 1 top = rstr.resample_to_grid(
2 sgrid, rstr.bands[0], method="min", extrapolate_edges=True
3 )
NameError: name 'rstr' is not defined
[9]:
fig, ax = plt.subplots(figsize=(7, 7))
pmv = PlotMapView(modelgrid=sgrid)
pc = pmv.plot_array(top)
pmv.plot_grid(lw=0.5)
plt.colorbar(pc, shrink=0.7);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[9], line 2
1 fig, ax = plt.subplots(figsize=(7, 7))
----> 2 pmv = PlotMapView(modelgrid=sgrid)
3 pc = pmv.plot_array(top)
4 pmv.plot_grid(lw=0.5)
NameError: name 'PlotMapView' is not defined
Vector (shapefile) intersection review
We can also intersect lines, points, and polygons with an existing modelgrid object. In this example, we’ll load up the watershed boundary and get the active and inactive areas of the grid. For more information on intersecting shapefiles with a modelgrid/model see this example
Our first step in this process is to get the watershed boundary. We can use the dataretrieval package to begin downloading information about the watershed from NWIS.
Note: this information has been stored in the repository as shapefiles if internet access is not available.
[10]:
# the get_info() call accepts a decimal lat/lon bounding box
griddf = sgrid.geo_dataframe.set_crs(epsg=26911)
grid_wgs = griddf.to_crs(epsg=4326)
wxmin, wxmax, wymin, wymax = grid_wgs.total_bounds
wgs_bounds = [wxmin, wymin, wxmax, wymax]
try:
info, metadata = nwis.get_info(bBox=[f"{i :.2f}" for i in wgs_bounds])
except (ValueError, ConnectionError, NameError):
info = gpd.read_file(geospatial_ws / "gage_info.shp")
info.head()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[10], line 2
1 # the get_info() call accepts a decimal lat/lon bounding box
----> 2 griddf = sgrid.geo_dataframe.set_crs(epsg=26911)
3 grid_wgs = griddf.to_crs(epsg=4326)
5 wxmin, wxmax, wymin, wymax = grid_wgs.total_bounds
NameError: name 'sgrid' is not defined
Reproject the gage data into UTM Zone 11 N
[11]:
info = info.to_crs(epsg=epsg)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[11], line 1
----> 1 info = info.to_crs(epsg=epsg)
NameError: name 'info' is not defined
And plot the gage data on the raster
[12]:
fig, ax = plt.subplots()
rstr.plot(ax=ax)
info.plot(
column="site_no",
ax=ax,
cmap="tab20",
legend=True,
legend_kwds={"fontsize": 6},
);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[12], line 3
1 fig, ax = plt.subplots()
----> 3 rstr.plot(ax=ax)
4 info.plot(
5 column="site_no",
6 ax=ax,
(...) 9 legend_kwds={"fontsize": 6},
10 );
NameError: name 'rstr' is not defined
Grab the site record for gage number 10343500
[13]:
sitedf = info.loc[info.site_no == "10343500"]
sitedf
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[13], line 1
----> 1 sitedf = info.loc[info.site_no == "10343500"]
2 sitedf
NameError: name 'info' is not defined
Get the upstream basin from nldi using the NWIS stream gage station code
[14]:
try:
basindf = nldi.get_basin(
feature_source="nwissite",
feature_id=f"USGS-{sitedf.site_no.values[0]}",
)
basindf = basindf.to_crs(epsg=epsg)
except (ValueError, ConnectionError, NameError):
basindf = gpd.read_file(geospatial_ws / "basin_boundary.shp")
---------------------------------------------------------------------------
DataSourceError Traceback (most recent call last)
Cell In[14], line 8
6 basindf = basindf.to_crs(epsg=epsg)
7 except (ValueError, ConnectionError, NameError):
----> 8 basindf = gpd.read_file(geospatial_ws / "basin_boundary.shp")
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/geopandas/io/file.py:316, in _read_file(filename, bbox, mask, columns, rows, engine, **kwargs)
313 filename = response.read()
315 if engine == "pyogrio":
--> 316 return _read_file_pyogrio(
317 filename, bbox=bbox, mask=mask, columns=columns, rows=rows, **kwargs
318 )
320 elif engine == "fiona":
321 if pd.api.types.is_file_like(filename):
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/geopandas/io/file.py:576, in _read_file_pyogrio(path_or_bytes, bbox, mask, rows, **kwargs)
567 warnings.warn(
568 "The 'include_fields' and 'ignore_fields' keywords are deprecated, and "
569 "will be removed in a future release. You can use the 'columns' keyword "
(...) 572 stacklevel=3,
573 )
574 kwargs["columns"] = kwargs.pop("include_fields")
--> 576 return pyogrio.read_dataframe(path_or_bytes, bbox=bbox, **kwargs)
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/pyogrio/geopandas.py:382, in read_dataframe(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, fid_as_index, use_arrow, on_invalid, arrow_to_pandas_kwargs, datetime_as_string, mixed_offsets_as_utc, **kwargs)
374 gdal_force_2d = False if use_arrow else force_2d
376 # Always read datetimes as string values to preserve (mixed) time zone info
377 # correctly. If arrow is not used, it is needed because numpy does not
378 # directly support time zones + performance is also a lot better. If arrow
379 # is used, needed because datetime columns don't support mixed time zone
380 # offsets + e.g. for .fgb files time zone info isn't handled correctly even
381 # for unique time zone offsets if datetimes are not read as string.
--> 382 result = read_func(
383 path_or_buffer,
384 layer=layer,
385 encoding=encoding,
386 columns=columns,
387 read_geometry=read_geometry,
388 force_2d=gdal_force_2d,
389 skip_features=skip_features,
390 max_features=max_features,
391 where=where,
392 bbox=bbox,
393 mask=mask,
394 fids=fids,
395 sql=sql,
396 sql_dialect=sql_dialect,
397 return_fids=fid_as_index,
398 datetime_as_string=True,
399 **kwargs,
400 )
402 if use_arrow:
403 import pyarrow as pa
File ~/micromamba/envs/pyclass-docs/lib/python3.11/site-packages/pyogrio/raw.py:200, in read(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, return_fids, datetime_as_string, **kwargs)
59 """Read OGR data source into numpy arrays.
60
61 IMPORTANT: non-linear geometry types (e.g., MultiSurface) are converted
(...) 196
197 """
198 dataset_kwargs = _preprocess_options_key_value(kwargs) if kwargs else {}
--> 200 return ogr_read(
201 get_vsi_path_or_buffer(path_or_buffer),
202 layer=layer,
203 encoding=encoding,
204 columns=columns,
205 read_geometry=read_geometry,
206 force_2d=force_2d,
207 skip_features=skip_features,
208 max_features=max_features or 0,
209 where=where,
210 bbox=bbox,
211 mask=_mask_to_wkb(mask),
212 fids=fids,
213 sql=sql,
214 sql_dialect=sql_dialect,
215 return_fids=return_fids,
216 dataset_kwargs=dataset_kwargs,
217 datetime_as_string=datetime_as_string,
218 )
File pyogrio/_io.pyx:1412, in pyogrio._io.ogr_read()
File pyogrio/_io.pyx:262, in pyogrio._io.ogr_open()
DataSourceError: data/unstructured_grid_data/basin_boundary.shp: No such file or directory
Plot up the basin boundary with the existing raster
[15]:
fig, ax = plt.subplots()
rstr.plot(ax=ax)
basindf.plot(ax=ax, facecolor="None", edgecolor="k", lw=1.5)
info.plot(
column="site_no",
ax=ax,
cmap="tab20",
legend=True,
legend_kwds={"fontsize": 6},
);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[15], line 2
1 fig, ax = plt.subplots()
----> 2 rstr.plot(ax=ax)
3 basindf.plot(ax=ax, facecolor="None", edgecolor="k", lw=1.5)
4 info.plot(
5 column="site_no",
6 ax=ax,
(...) 9 legend_kwds={"fontsize": 6},
10 );
NameError: name 'rstr' is not defined
Doing the intersection
[16]:
ix = GridIntersect(sgrid, method="vertex")
result = ix.intersect(basindf.geometry.values[0], contains_centroid=True)
rowcol = result["cellids"]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[16], line 1
----> 1 ix = GridIntersect(sgrid, method="vertex")
2 result = ix.intersect(basindf.geometry.values[0], contains_centroid=True)
3 rowcol = result["cellids"]
NameError: name 'GridIntersect' is not defined
[17]:
row, col = list(zip(*rowcol))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[17], line 1
----> 1 row, col = list(zip(*rowcol))
NameError: name 'rowcol' is not defined
Building idomain from the intersection
[18]:
idomain = np.zeros((nlay, nrow, ncol), dtype=int)
idomain[:, row, col] = 1
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[18], line 1
----> 1 idomain = np.zeros((nlay, nrow, ncol), dtype=int)
2 idomain[:, row, col] = 1
NameError: name 'nlay' is not defined
Putting it all together into a completed StructuredGrid
[19]:
sgrid = StructuredGrid(
delc=sgrid.delc,
delr=sgrid.delr,
top=top,
botm=sgrid.botm,
idomain=idomain,
nlay=sgrid.nlay,
nrow=sgrid.nrow,
ncol=sgrid.ncol,
xoff=sgrid.xoffset,
yoff=sgrid.yoffset,
crs=sgrid.crs,
)
sgrid
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[19], line 1
----> 1 sgrid = StructuredGrid(
2 delc=sgrid.delc,
3 delr=sgrid.delr,
4 top=top,
5 botm=sgrid.botm,
6 idomain=idomain,
7 nlay=sgrid.nlay,
8 nrow=sgrid.nrow,
9 ncol=sgrid.ncol,
10 xoff=sgrid.xoffset,
11 yoff=sgrid.yoffset,
12 crs=sgrid.crs,
13 )
14 sgrid
NameError: name 'StructuredGrid' is not defined
Plotting the completed grid with gage locations and the watershed boundary
[20]:
fig, ax = plt.subplots(figsize=(5, 5))
pmv = PlotMapView(modelgrid=sgrid)
pc = pmv.plot_array(sgrid.top)
ib = pmv.plot_inactive()
pmv.plot_grid(lw=0.5)
plt.colorbar(pc, shrink=0.7)
basindf.plot(ax=ax, facecolor="None", edgecolor="r", lw=1.5)
sitedf.plot(
column="site_no",
ax=ax,
cmap="tab20",
legend=True,
legend_kwds={"fontsize": 6},
zorder=6
);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[20], line 2
1 fig, ax = plt.subplots(figsize=(5, 5))
----> 2 pmv = PlotMapView(modelgrid=sgrid)
3 pc = pmv.plot_array(sgrid.top)
4 ib = pmv.plot_inactive()
NameError: name 'PlotMapView' is not defined
Because this is a watershed, we can also overlay a shapefile of the streams and get the stream cells using GridIntersect
NHD-Plus stream lines can be downloaded using streamgage information with the dataretrieval package
[21]:
try:
nhddf = nldi.get_flowlines(
navigation_mode="UT",
distance=999,
feature_source="nwissite",
feature_id=f"USGS-{sitedf.site_no.values[0]}",
)
nhddf = nhddf.to_crs(epsg=epsg)
except (ValueError, ConnectionError, NameError):
nhddf = gpd.read_file(geospatial_ws / "nhd_streams.shp")
nhddf.head()
[21]:
| nhdplus_co | geometry | |
|---|---|---|
| 0 | 8933522 | LINESTRING (221038.59 4369598.73, 221076.943 4... |
| 1 | 8933608 | LINESTRING (220451.353 4367186.202, 220542.627... |
| 2 | 8933524 | LINESTRING (220541.209 4369697.393, 220691.816... |
| 3 | 8934342 | LINESTRING (219823.023 4371753.116, 219905.68 ... |
| 4 | 8933520 | LINESTRING (220240.836 4369758.678, 220245.466... |
Intersect stream lines with the existing modelgrid
[22]:
rowcols = []
ix = GridIntersect(sgrid, method="vertex")
for geom in nhddf.geometry.values:
rcs = ix.intersects(geom)["cellids"]
rowcols.extend(list(rcs))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[22], line 2
1 rowcols = []
----> 2 ix = GridIntersect(sgrid, method="vertex")
3 for geom in nhddf.geometry.values:
4 rcs = ix.intersects(geom)["cellids"]
NameError: name 'GridIntersect' is not defined
[23]:
row, col = list(zip(*rowcols))
strms = np.zeros((nrow, ncol))
strms[row, col] = 1
strms
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[23], line 1
----> 1 row, col = list(zip(*rowcols))
2 strms = np.zeros((nrow, ncol))
3 strms[row, col] = 1
ValueError: not enough values to unpack (expected 2, got 0)
Plot the intersection results
[24]:
fig, ax = plt.subplots(figsize=(5, 5))
pmv = PlotMapView(modelgrid=sgrid, ax=ax)
pc = pmv.plot_array(sgrid.top, cmap="viridis")
pmv.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.5)
plt.colorbar(pc, shrink=0.7)
nhddf.plot(ax=ax);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[24], line 2
1 fig, ax = plt.subplots(figsize=(5, 5))
----> 2 pmv = PlotMapView(modelgrid=sgrid, ax=ax)
3 pc = pmv.plot_array(sgrid.top, cmap="viridis")
4 pmv.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
NameError: name 'PlotMapView' is not defined
Information from the grid we’ve created here can now be used for generating more complex meshes.
Irregular structured Grid (DIS)
In this example, an irregular structured grid is created to demonstrate how to add local refinement to an area of the model using a structured (rectilinear) grid.
[25]:
lx = xmax - xmin
ly = ymax - ymin
cellsize = 200
rcellsize = 100
smooth = np.linspace(100, 200, 10).tolist()
delr = 9 * [cellsize] + smooth[::-1] + 8 * [rcellsize] + smooth + 9 * [cellsize]
delc = 8 * [cellsize] + smooth[::-1] + 7 * [rcellsize] + smooth + 8 * [cellsize]
nlay = 1
nrow = len(delc)
ncol = len(delr)
# create some fake data for the moment
top = np.ones((nrow, ncol))
botm = np.zeros((nlay, nrow, ncol))
idomain = np.ones(botm.shape, dtype=int)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[25], line 1
----> 1 lx = xmax - xmin
2 ly = ymax - ymin
3 cellsize = 200
NameError: name 'xmax' is not defined
[26]:
rfgrid = StructuredGrid(
delc=np.array(delc),
delr=np.array(delr),
nlay=nlay,
top=top,
botm=botm,
idomain=idomain,
xoff=xmin,
yoff=ymin
)
print(nlay, nrow, ncol)
print(rfgrid.nrow, rfgrid.ncol)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[26], line 1
----> 1 rfgrid = StructuredGrid(
2 delc=np.array(delc),
3 delr=np.array(delr),
4 nlay=nlay,
5 top=top,
6 botm=botm,
7 idomain=idomain,
8 xoff=xmin,
9 yoff=ymin
10 )
11 print(nlay, nrow, ncol)
12 print(rfgrid.nrow, rfgrid.ncol)
NameError: name 'StructuredGrid' is not defined
Intersect the basin boundary with the grid to set active and inactive cells (idomain)
[27]:
ix = GridIntersect(rfgrid, method="vertex")
result = ix.intersect(basindf.geometry.values[0], contains_centroid=True)
rowcol = result["cellids"]
row, col = list(zip(*rowcol))
idomain = np.zeros((nlay, nrow, ncol), dtype=int)
idomain[:, row, col] = 1
# tricky way so we don't have to recreate the grid
rfgrid._idomain = idomain
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[27], line 1
----> 1 ix = GridIntersect(rfgrid, method="vertex")
2 result = ix.intersect(basindf.geometry.values[0], contains_centroid=True)
3 rowcol = result["cellids"]
NameError: name 'GridIntersect' is not defined
Resample the top elevation
[28]:
top = rstr.resample_to_grid(
rfgrid, rstr.bands[0], method="min", extrapolate_edges=True
)
# tricky way so we don't have to recreate the grid
rfgrid._top = top
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[28], line 1
----> 1 top = rstr.resample_to_grid(
2 rfgrid, rstr.bands[0], method="min", extrapolate_edges=True
3 )
5 # tricky way so we don't have to recreate the grid
6 rfgrid._top = top
NameError: name 'rstr' is not defined
Now we can plot the grid
[29]:
fig, ax = plt.subplots(figsize=(5, 5))
pmv = PlotMapView(modelgrid=rfgrid)
pc = pmv.plot_array(rfgrid.top, cmap="viridis")
ib = pmv.plot_inactive()
pmv.plot_grid(lw=0.5)
plt.colorbar(pc, shrink=0.7)
basindf.plot(ax=ax, facecolor="None", edgecolor="r", lw=1.5)
sitedf.plot(
column="site_no",
ax=ax,
cmap="tab20",
legend=True,
legend_kwds={"fontsize": 6},
zorder=6
);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[29], line 2
1 fig, ax = plt.subplots(figsize=(5, 5))
----> 2 pmv = PlotMapView(modelgrid=rfgrid)
3 pc = pmv.plot_array(rfgrid.top, cmap="viridis")
4 ib = pmv.plot_inactive()
NameError: name 'PlotMapView' is not defined
Local Grid Refinement (LGR) mesh
A Local Grid Refinement (LGR) mesh can be created in an area of interest to get finer discretization than the existing/surrounding mesh. FloPy’s Lgr utility, creates the refined mesh as a seperate modelgrid, and calculates the exchanges between the multi-model simulation mesh that results from refinement.
The Lgr utility has a number of input parameters:
nlayp: the number of parent model layersnrowp: the number of parent model rowsncolp: the number of parent model columnsdelrp: the parent model delr arraydelcp: the parent model delc arraytopp: the parent model top arraybotmp: the parent model botm arrayidomainp: an idomain array that is used to create the child grid. Values of 1 indicate parent model cells, values of 0 indicate child model cells. The child model must be a rectangular region that is continuous.ncpp: number of child cells along the face of a parent cellncppl: number of child cells per parent layerxllp: (optional) x location of parent grid lower left corneryllp: (optional) y-location of parent grid lower left corner
Load up the refinement area polygon
[30]:
lgr_gdf = gpd.read_file(geospatial_ws / "lgr_bound.shp")
Plot it to see if it lines up properly with our model
[31]:
fig, ax = plt.subplots(figsize=(5, 5))
pmv = PlotMapView(modelgrid=sgrid, ax=ax)
pc = pmv.plot_array(sgrid.top, cmap="viridis")
pmv.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.5)
nhddf.plot(ax=ax)
lgr_gdf.plot(ax=ax, alpha=0.5)
plt.colorbar(pc, shrink=0.7);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[31], line 2
1 fig, ax = plt.subplots(figsize=(5, 5))
----> 2 pmv = PlotMapView(modelgrid=sgrid, ax=ax)
3 pc = pmv.plot_array(sgrid.top, cmap="viridis")
4 pmv.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
NameError: name 'PlotMapView' is not defined
Creating the parent/child idomainp array for Lgr using GridIntersect
[32]:
lgr_gdf
[32]:
| Id | geometry | |
|---|---|---|
| 0 | 0 | POLYGON ((221180.716 4369310.065, 219973.134 4... |
Activity: Create the idomainp (parent, child) array from the polygon provided by the lgr geodataframe
idomainp should be the same shape as the existing modelgrid’s idomain array. The child model extent should be represented with the value 0 and the parent 1.
Hint: GridIntersect() can be used to find the cellids where the lgr polygon intersects with the existing modelgrid
[33]:
# live code this/give it as an activity
idomainp = np.ones(sgrid.idomain.shape, dtype=int)
gix = GridIntersect(sgrid, method="vertex")
result = gix.intersect(lgr_gdf.geometry.values[0])
cid = result["cellids"]
crow, ccol = list(zip(*cid))
idomainp[:, crow, ccol] = 0
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[33], line 2
1 # live code this/give it as an activity
----> 2 idomainp = np.ones(sgrid.idomain.shape, dtype=int)
4 gix = GridIntersect(sgrid, method="vertex")
5 result = gix.intersect(lgr_gdf.geometry.values[0])
NameError: name 'sgrid' is not defined
Creating the Lgr object/child grid
[34]:
lgr = Lgr(
nlayp=sgrid.nlay,
nrowp=sgrid.nrow,
ncolp=sgrid.ncol,
delrp=sgrid.delr,
delcp=sgrid.delc,
topp=sgrid.top,
botmp=sgrid.botm,
idomainp=idomainp,
ncpp=4,
xllp=sgrid.xoffset,
yllp=sgrid.yoffset,
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[34], line 1
----> 1 lgr = Lgr(
2 nlayp=sgrid.nlay,
3 nrowp=sgrid.nrow,
4 ncolp=sgrid.ncol,
5 delrp=sgrid.delr,
6 delcp=sgrid.delc,
7 topp=sgrid.top,
8 botmp=sgrid.botm,
9 idomainp=idomainp,
10 ncpp=4,
11 xllp=sgrid.xoffset,
12 yllp=sgrid.yoffset,
13 )
NameError: name 'Lgr' is not defined
Get a StructuredGrid of the child model
[35]:
childgrid = lgr.child.modelgrid
childgrid
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[35], line 1
----> 1 childgrid = lgr.child.modelgrid
2 childgrid
NameError: name 'lgr' is not defined
Resample top elevations to the child grid
[36]:
childtop = rstr.resample_to_grid(childgrid, band=rstr.bands[0], method="min")
print(childtop.shape)
print(childgrid.shape)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[36], line 1
----> 1 childtop = rstr.resample_to_grid(childgrid, band=rstr.bands[0], method="min")
2 print(childtop.shape)
3 print(childgrid.shape)
NameError: name 'rstr' is not defined
Reset the top elevations in the childgrid object
[37]:
childgrid._top = childtop
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[37], line 1
----> 1 childgrid._top = childtop
NameError: name 'childtop' is not defined
Intersect streams with the Child mesh to define stream cells
[38]:
ix = GridIntersect(childgrid, method="vertex")
cstrmcells = []
for geom in nhddf.geometry.values:
rcs = ix.intersects(geom)["cellids"]
if len(rowcol) > 0:
cstrmcells.extend(list(rcs))
cstrms = np.zeros((childgrid.nrow, childgrid.ncol), dtype=int)
row, col = list(zip(*cstrmcells))
cstrms[row, col] = 1
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[38], line 1
----> 1 ix = GridIntersect(childgrid, method="vertex")
2 cstrmcells = []
3 for geom in nhddf.geometry.values:
NameError: name 'GridIntersect' is not defined
And now we can visualize the new LGR mesh
[39]:
fig, ax = plt.subplots(figsize=(10, 10))
vmin, vmax = np.min(sgrid.top), np.max(sgrid.top)
# plot the parent
pmvp = PlotMapView(modelgrid=sgrid, ax=ax)
ptop = sgrid.top.copy()
ptop[crow, ccol] = np.nan
strms[crow, ccol] = np.nan
pc = pmvp.plot_array(ptop, cmap="viridis", vmin=vmin, vmax=vmax)
pmvp.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmvp.plot_inactive()
pmvp.plot_grid(lw=0.3)
nhddf.plot(ax=ax)
# plot the child
pmvc = PlotMapView(modelgrid=childgrid, ax=ax, extent=sgrid.extent)
pmvc.plot_array(childtop, cmap="viridis", vmin=vmin, vmax=vmax)
pmvc.plot_array(cstrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmvc.plot_grid(lw=0.3)
plt.colorbar(pc, shrink=0.7);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[39], line 2
1 fig, ax = plt.subplots(figsize=(10, 10))
----> 2 vmin, vmax = np.min(sgrid.top), np.max(sgrid.top)
4 # plot the parent
5 pmvp = PlotMapView(modelgrid=sgrid, ax=ax)
NameError: name 'sgrid' is not defined
Quadtree Mesh
A quadtree mesh is an unstructured refinement to a rectilinear modelgrid. In a quadtree mesh each node is split into 4 child nodes per level of refinement. The figure below is from Lien and others, 2019 and shows multiple levels of recursive refinement.
For this example, we will be using GRIDGEN and the FloPy utility Gridgen to create quadtree refinement around the streams in our example.
[40]:
gridgen_dir = data_ws / "sagehen_gridgen"
gridgen_dir.mkdir(exist_ok=True)
The Gridgen class a number of input parameters and further documentation/examples of the class can be found here. For our example, we will be refining the area around the stream channels.
[41]:
g = Gridgen(sgrid, model_ws=gridgen_dir)
for geom in nhddf.geometry.values:
xy = list(zip(*geom.coords.xy))
g.add_refinement_features([xy,], "line", 2, [0,])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[41], line 1
----> 1 g = Gridgen(sgrid, model_ws=gridgen_dir)
2 for geom in nhddf.geometry.values:
3 xy = list(zip(*geom.coords.xy))
NameError: name 'Gridgen' is not defined
[42]:
g.build()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[42], line 1
----> 1 g.build()
NameError: name 'g' is not defined
After gridgen builds the mesh, it can be used to create a VertexGrid (or DISV) that represents the basin
[43]:
gridprops = g.get_gridprops_disv()
gridprops.pop("nvert")
quadgrid = VertexGrid(**gridprops)
quadgrid.extent
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[43], line 1
----> 1 gridprops = g.get_gridprops_disv()
2 gridprops.pop("nvert")
4 quadgrid = VertexGrid(**gridprops)
NameError: name 'g' is not defined
Resample top elevations to the quadtree mesh and intersect the basin boundary and stream vectors
[44]:
# create the top array
top = rstr.resample_to_grid(quadgrid, band=rstr.bands[0], method="min")
quadgrid._top = top
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[44], line 2
1 # create the top array
----> 2 top = rstr.resample_to_grid(quadgrid, band=rstr.bands[0], method="min")
3 quadgrid._top = top
NameError: name 'rstr' is not defined
[45]:
# create the idomain array
idomain = np.zeros(quadgrid.shape, dtype=int)
ix = GridIntersect(quadgrid, method="vertex")
nodes = ix.intersect(basindf.geometry.values[0], contains_centroid=True)["cellids"]
idomain[:, list(nodes)] = 1
quadgrid._idomain = idomain
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[45], line 2
1 # create the idomain array
----> 2 idomain = np.zeros(quadgrid.shape, dtype=int)
3 ix = GridIntersect(quadgrid, method="vertex")
4 nodes = ix.intersect(basindf.geometry.values[0], contains_centroid=True)["cellids"]
NameError: name 'quadgrid' is not defined
[46]:
# create an array of stream cells
qstr = []
ix = GridIntersect(quadgrid, method="vertex")
for geom in nhddf.geometry.values:
nodes = ix.intersects(geom)["cellids"]
qstr.extend(list(nodes))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[46], line 3
1 # create an array of stream cells
2 qstr = []
----> 3 ix = GridIntersect(quadgrid, method="vertex")
4 for geom in nhddf.geometry.values:
5 nodes = ix.intersects(geom)["cellids"]
NameError: name 'GridIntersect' is not defined
[47]:
qstrms = np.zeros((quadgrid.ncpl,), dtype=int)
qstrms[qstr] = 1
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[47], line 1
----> 1 qstrms = np.zeros((quadgrid.ncpl,), dtype=int)
2 qstrms[qstr] = 1
NameError: name 'quadgrid' is not defined
Plot the quadtree mesh with top elevations and streams
[48]:
fig, ax = plt.subplots(figsize=(5, 5))
pmv = PlotMapView(modelgrid=quadgrid)
pc = pmv.plot_array(quadgrid.top)
pmv.plot_array(qstrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=ax)
plt.colorbar(pc, shrink=0.7);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[48], line 3
1 fig, ax = plt.subplots(figsize=(5, 5))
----> 3 pmv = PlotMapView(modelgrid=quadgrid)
4 pc = pmv.plot_array(quadgrid.top)
5 pmv.plot_array(qstrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
NameError: name 'PlotMapView' is not defined
Triangular Mesh
An unstructured/vertex based triangular mesh can be generated using the code “triangle”(Shewchuk, 2002). And the FloPy utility, Triangle that creates inputs for and translates the outputs from “triangle”.
For this example, we will be using triangle to generate a mesh and add refinement around the streams.
[49]:
tri_dir = data_ws / "sagehen_tri"
tri_dir.mkdir(exist_ok=True)
For this example, we will use the watershed boundary we downloaded from NLDI and create a dissovled polygon of stream segments to generate our triangulation regions.
[50]:
basindf.head()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[50], line 1
----> 1 basindf.head()
NameError: name 'basindf' is not defined
We will also dissolve and buffer the stream network into a single polygon that defines our area of refinement
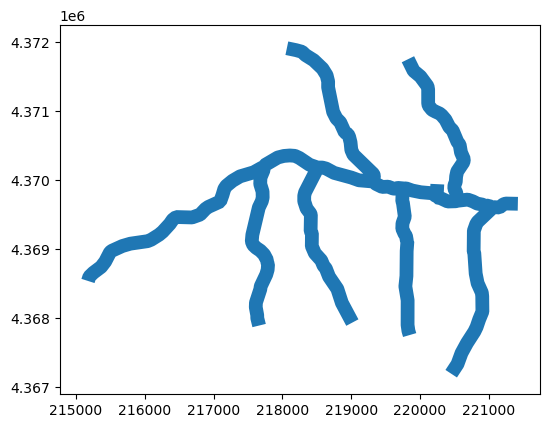
[51]:
bufgdf = nhddf.dissolve()
bufgdf["geometry"] = bufgdf.geometry.buffer(100, cap_style=2, join_style=3)
bufgdf.plot();
These two polygons can now be used to generate a triangular mesh with the Triangle utility. The Triangle class has a number of input parameters. For more detail on the inputs and additional examples please see this notebook
[52]:
# define point locations within the watershed and the stream for adding regions
wsloc = (220000, 4368000)
stloc = (219250, 4370000)
[53]:
tri = Triangle(angle=30, model_ws=tri_dir)
# define the model/mesh boundary
tri.add_polygon(basindf.geometry.values[0])
tri.add_region(wsloc, 0, maximum_area=100 * 300)
# define the stream refinement area
tri.add_polygon(bufgdf.geometry.values[0])
tri.add_region(stloc, 1, maximum_area=40 * 40)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[53], line 1
----> 1 tri = Triangle(angle=30, model_ws=tri_dir)
3 # define the model/mesh boundary
4 tri.add_polygon(basindf.geometry.values[0])
NameError: name 'Triangle' is not defined
[54]:
tri.build()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[54], line 1
----> 1 tri.build()
NameError: name 'tri' is not defined
Visualize the triangular mesh
[55]:
fig, ax = plt.subplots(figsize=(6, 6))
tri.plot(ax=ax)
nhddf.plot(ax=ax);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[55], line 2
1 fig, ax = plt.subplots(figsize=(6, 6))
----> 2 tri.plot(ax=ax)
3 nhddf.plot(ax=ax);
NameError: name 'tri' is not defined
Create a VertexGrid from the triangular mesh and resample the land surface elevations
[56]:
cell2d = tri.get_cell2d()
vertices = tri.get_vertices()
ncpl = len(cell2d)
nlay = 1
idomain = np.ones((nlay, ncpl), dtype=int)
# set fake values for top and botm for now)
top = np.ones((ncpl,))
botm = np.zeros((nlay, ncpl))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[56], line 1
----> 1 cell2d = tri.get_cell2d()
2 vertices = tri.get_vertices()
3 ncpl = len(cell2d)
NameError: name 'tri' is not defined
[57]:
trigrid = VertexGrid(
vertices=vertices,
cell2d=cell2d,
ncpl=ncpl,
nlay=nlay,
idomain=idomain,
top=top,
botm=botm,
crs="EPSG:26911",
)
trigrid.extent
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[57], line 1
----> 1 trigrid = VertexGrid(
2 vertices=vertices,
3 cell2d=cell2d,
4 ncpl=ncpl,
5 nlay=nlay,
6 idomain=idomain,
7 top=top,
8 botm=botm,
9 crs="EPSG:26911",
10 )
11 trigrid.extent
NameError: name 'VertexGrid' is not defined
[58]:
top = rstr.resample_to_grid(
trigrid, band=rstr.bands[0], method="min", extrapolate_edges=True
)
trigrid._top = top
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[58], line 1
----> 1 top = rstr.resample_to_grid(
2 trigrid, band=rstr.bands[0], method="min", extrapolate_edges=True
3 )
4 trigrid._top = top
NameError: name 'rstr' is not defined
And perform intersection to identify stream cells
[59]:
tristr = []
ix = GridIntersect(trigrid, method="vertex")
for geom in nhddf.geometry.values:
nodes = ix.intersects(geom)["cellids"]
tristr.extend(list(nodes))
tristrms = np.zeros((trigrid.ncpl,), dtype=int)
tristrms[tristr] = 1
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[59], line 2
1 tristr = []
----> 2 ix = GridIntersect(trigrid, method="vertex")
3 for geom in nhddf.geometry.values:
4 nodes = ix.intersects(geom)["cellids"]
NameError: name 'GridIntersect' is not defined
Now to plot up the triangular grid
[60]:
fig, ax = plt.subplots(figsize=(7, 7))
pmv = PlotMapView(modelgrid=trigrid)
pc = pmv.plot_array(trigrid.top)
pmv.plot_array(tristrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=ax)
plt.colorbar(pc, shrink=0.7);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[60], line 3
1 fig, ax = plt.subplots(figsize=(7, 7))
----> 3 pmv = PlotMapView(modelgrid=trigrid)
4 pc = pmv.plot_array(trigrid.top)
5 pmv.plot_array(tristrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
NameError: name 'PlotMapView' is not defined
Voronoi Mesh
A voronoi mesh can be generated from the circumcirle centroids of each triangular node in a Delaunay triangulation. The centroid of each circumcirle produces a vertex for the voronoi mesh and these vertices are connected via adjacency relationships of the triangulation.
FloPy has a built in utility VoronoiGrid that produces voronoi meshes from Delaunay triangulations. For more information and additional examples on VoronoiGrid’s usage, see this notebook
To generate a voronoi mesh, we just need to pass our triangluation object (tri) to the VoronoiGrid utility
[61]:
vor = VoronoiGrid(tri)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[61], line 1
----> 1 vor = VoronoiGrid(tri)
NameError: name 'VoronoiGrid' is not defined
Build a VertexGrid
[62]:
gridprops = vor.get_gridprops_vertexgrid()
nlay = 1
idomain = np.ones(gridprops["ncpl"], dtype=int)
top = idomain.copy().astype(float)
botm = np.zeros((nlay, gridprops["ncpl"]))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[62], line 1
----> 1 gridprops = vor.get_gridprops_vertexgrid()
2 nlay = 1
3 idomain = np.ones(gridprops["ncpl"], dtype=int)
NameError: name 'vor' is not defined
[63]:
vorgrid = VertexGrid(
nlay=nlay, idomain=idomain, top=top, botm=botm, **gridprops
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[63], line 1
----> 1 vorgrid = VertexGrid(
2 nlay=nlay, idomain=idomain, top=top, botm=botm, **gridprops
3 )
NameError: name 'VertexGrid' is not defined
Resample the land surface elevation from the DEM to the voronoi grid
[64]:
top = rstr.resample_to_grid(
vorgrid, band=rstr.bands[0], method="min", extrapolate_edges=True
)
vorgrid._top = top
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[64], line 1
----> 1 top = rstr.resample_to_grid(
2 vorgrid, band=rstr.bands[0], method="min", extrapolate_edges=True
3 )
4 vorgrid._top = top
NameError: name 'rstr' is not defined
Identify stream cells with GridIntersect
[65]:
vorst = []
ix = GridIntersect(vorgrid, method="vertex")
for geom in nhddf.geometry.values:
nodes = ix.intersects(geom)["cellids"]
vorst.extend(list(nodes))
vorstrms = np.zeros((vorgrid.ncpl,), dtype=int)
vorstrms[vorst] = 1
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[65], line 2
1 vorst = []
----> 2 ix = GridIntersect(vorgrid, method="vertex")
3 for geom in nhddf.geometry.values:
4 nodes = ix.intersects(geom)["cellids"]
NameError: name 'GridIntersect' is not defined
Finally plot the voronoi mesh with the DEM data and stream locations
[66]:
fig, ax = plt.subplots(figsize=(7, 7))
pmv = PlotMapView(modelgrid=vorgrid)
pc = pmv.plot_array(vorgrid.top)
pmv.plot_array(vorstrms, masked_values=[0], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=ax)
plt.colorbar(pc, shrink=0.7);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[66], line 3
1 fig, ax = plt.subplots(figsize=(7, 7))
----> 3 pmv = PlotMapView(modelgrid=vorgrid)
4 pc = pmv.plot_array(vorgrid.top)
5 pmv.plot_array(vorstrms, masked_values=[0], alpha=0.3, cmap="Reds_r")
NameError: name 'PlotMapView' is not defined
plot the meshes we produced side by side for comparison!
[67]:
from flopy.plot import styles
from matplotlib.gridspec import GridSpec
with styles.USGSMap():
fig, axm = plt.subplots(nrows=3, ncols=2, figsize=(10,12))
axs = axm.ravel()
# structured grid
pmv = PlotMapView(modelgrid=sgrid, ax=axs[0])
pmv.plot_array(sgrid.top, cmap="viridis")
pmv.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=axs[0])
styles.heading(axs[0], "A. ", heading="Structured Grid", x=0.08)
# non-uniform DIS
pmv = PlotMapView(modelgrid=rfgrid, ax=axs[1])
pmv.plot_array(rfgrid.top, cmap="viridis")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=axs[1])
styles.heading(axs[1], "B.", heading="Structured Grid: non-uniform", x=0.08)
# LGR grid
pmvp = PlotMapView(modelgrid=sgrid, ax=axs[2])
ptop = sgrid.top.copy()
ptop[crow, ccol] = np.nan
strms[crow, ccol] = np.nan
pc = pmvp.plot_array(ptop, cmap="viridis", vmin=vmin, vmax=vmax)
pmvp.plot_array(strms, masked_values=[0], alpha=0.3, cmap="Reds_r")
pmvp.plot_inactive()
pmvp.plot_grid(lw=0.3)
nhddf.plot(ax=axs[2])
pmvc = PlotMapView(modelgrid=childgrid, ax=axs[2])
pmvc.plot_array(childtop, cmap="viridis", vmin=vmin, vmax=vmax)
pmvc.plot_array(cstrms, masked_values=[0], alpha=0.3, cmap="Reds_r")
pmvc.plot_grid(lw=0.3)
styles.heading(axs[2], "C. ", heading="LGR Grid", x=0.08)
# Quadtree grid
pmv = PlotMapView(modelgrid=quadgrid, ax=axs[3])
pc = pmv.plot_array(quadgrid.top)
pmv.plot_array(qstrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.2)
nhddf.plot(ax=axs[3])
styles.heading(axs[3], "D. ", heading="Quadtree Grid", x=0.08)
# Triangular grid
pmv = PlotMapView(modelgrid=trigrid, ax=axs[4])
pc = pmv.plot_array(trigrid.top)
pmv.plot_array(tristrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=axs[4])
styles.heading(axs[4], "E. ", heading="Triangular Grid", x=0.08)
# Voronoi grid
pmv = PlotMapView(modelgrid=vorgrid, ax=axs[5])
pc = pmv.plot_array(vorgrid.top)
pmv.plot_array(vorstrms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
pmv.plot_inactive()
pmv.plot_grid(lw=0.3)
nhddf.plot(ax=axs[5])
styles.heading(axs[5], "F. ", heading="Voronoi Grid", x=0.08)
# colorbar
# ax5.spines[["left", "right", "top", "bottom"]].set_visible(False)
fig.colorbar(
pc,
ax=axm,
location="top",
label="Land surface elevation, in meters",
shrink=0.5,
fraction=.05
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[67], line 9
6 axs = axm.ravel()
8 # structured grid
----> 9 pmv = PlotMapView(modelgrid=sgrid, ax=axs[0])
10 pmv.plot_array(sgrid.top, cmap="viridis")
11 pmv.plot_array(strms, masked_values=[0,], alpha=0.3, cmap="Reds_r")
NameError: name 'PlotMapView' is not defined
More information
For more inforation and and examples on how to use the utilities presented in this notebook please see:
Examples
API reference
Activity (If time permits); Build a Voronoi mesh/Vertex Grid:
Define a voronoi grid for the basin upstream of the Black Earth Creek streamgauge near Cross Plains, WI. The upstream basin information is supplied by NLDI and has been saved to the class repo in case of internet connectivity or NLDI service issues.
Some of the previous code can be re-used to create a voronoi mesh and subsequent VertexGrid representation of the basin. See the Triangular and Voronoi grid sections of the notebook.
Optional, use the Upstream Main segment of Black Earth Creek to refine the voronoi mesh around the stream location. Refinement control points have been pre-defined below.
[68]:
epsg = 5070 # NAD83 CONUS Albers
# try:
# todo: update this for black earth creek near cross plains WI site-no 05406479
be_cr_nr_cp = "05406479"
basindf = nldi.get_basin(
feature_source="nwissite", feature_id=f"USGS-{be_cr_nr_cp}"
)
basindf = basindf.to_crs(epsg=epsg)
basindf.to_file(geospatial_ws / "black_earth_basin.shp")
# except (ValueError, ConnectionError, NameError):
# basindf = gpd.read_file(geospatial_ws / "black_earth_basin.shp")
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[68], line 5
2 # try:
3 # todo: update this for black earth creek near cross plains WI site-no 05406479
4 be_cr_nr_cp = "05406479"
----> 5 basindf = nldi.get_basin(
6 feature_source="nwissite", feature_id=f"USGS-{be_cr_nr_cp}"
7 )
8 basindf = basindf.to_crs(epsg=epsg)
9 basindf.to_file(geospatial_ws / "black_earth_basin.shp")
NameError: name 'nldi' is not defined
[69]:
try:
nhddf = nldi.get_flowlines(
navigation_mode="UM",
distance=9999,
feature_source="nwissite",
feature_id=f"USGS-{be_cr_nr_cp}",
)
nhddf = nhddf.to_crs(epsg=epsg)
nhddf.to_file(geospatial_ws / "black_earth_creek_main.shp")
except (ValueError, ConnectionError, NameError):
nhddf = gpd.read_file(geospatial_ws / "black_earth_creek_main.shp")
Visualize the example “study area”
[70]:
fig, ax = plt.subplots()
basindf.plot(ax=ax, facecolor="None", edgecolor="k")
nhddf.plot(ax=ax)
ctx.add_basemap(
ax=ax,
crs=basindf.crs,
source=ctx.providers.USGS.USTopo
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[70], line 3
1 fig, ax = plt.subplots()
----> 3 basindf.plot(ax=ax, facecolor="None", edgecolor="k")
4 nhddf.plot(ax=ax)
5 ctx.add_basemap(
6 ax=ax,
7 crs=basindf.crs,
8 source=ctx.providers.USGS.USTopo
9 )
NameError: name 'basindf' is not defined
Grid refinement control points are defined here for the optional part of this exercise
[71]:
wsloc = basindf.centroid.values[0]
stloc = list(zip(*nhddf.geometry.values[10].xy))[0]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[71], line 1
----> 1 wsloc = basindf.centroid.values[0]
2 stloc = list(zip(*nhddf.geometry.values[10].xy))[0]
NameError: name 'basindf' is not defined
[ ]:
[ ]:
[ ]: